Genome-Wide Gene Expression Analysis Shows AKAP13-Mediated PKD1 Signaling Regulates the Transcriptional Response to Cardiac Hypertrophy
- PMID: 26192751
- PMCID: PMC4508115
- DOI: 10.1371/journal.pone.0132474
Genome-Wide Gene Expression Analysis Shows AKAP13-Mediated PKD1 Signaling Regulates the Transcriptional Response to Cardiac Hypertrophy
Abstract
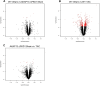
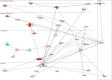
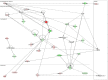
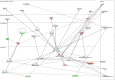
In the heart, scaffolding proteins such as A-Kinase Anchoring Proteins (AKAPs) play a crucial role in normal cellular function by serving as a signaling hub for multiple protein kinases including protein kinase D1 (PKD1). Under cardiac hypertrophic conditions AKAP13 anchored PKD1 activates the transcription factor MEF2 leading to subsequent fetal gene activation and hypertrophic response. We used an expression microarray to identify the global transcriptional response in the hearts of wild-type mice expressing the native form of AKAP13 compared to a gene-trap mouse model expressing a truncated form of AKAP13 that is unable to bind PKD1 (AKAP13-ΔPKD1). Microarray analysis showed that AKAP13-ΔPKD1 mice broadly failed to exhibit the transcriptional profile normally associated with compensatory cardiac hypertrophy following trans-aortic constriction (TAC). The identified differentially expressed genes in WT and AKAP13-ΔPKD1 hearts are vital for the compensatory hypertrophic response to pressure-overload and include myofilament, apoptotic, and cell growth/differentiation genes in addition to genes not previously identified as affected by AKAP13-anchored PKD1. Our results show that AKAP13-PKD1 signaling is critical for transcriptional regulation of key contractile, cell death, and metabolic pathways during the development of compensatory hypertrophy in vivo.
Conflict of interest statement
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

