Paired exome analysis of Barrett's esophagus and adenocarcinoma
- PMID: 26192918
- PMCID: PMC4552571
- DOI: 10.1038/ng.3343
Paired exome analysis of Barrett's esophagus and adenocarcinoma
Abstract
Barrett's esophagus is thought to progress to esophageal adenocarcinoma (EAC) through a stepwise progression with loss of CDKN2A followed by TP53 inactivation and aneuploidy. Here we present whole-exome sequencing from 25 pairs of EAC and Barrett's esophagus and from 5 patients whose Barrett's esophagus and tumor were extensively sampled. Our analysis showed that oncogene amplification typically occurred as a late event and that TP53 mutations often occurred early in Barrett's esophagus progression, including in non-dysplastic epithelium. Reanalysis of additional EAC exome data showed that the majority (62.5%) of EACs emerged following genome doubling and that tumors with genomic doubling had different patterns of genomic alterations, with more frequent oncogenic amplification and less frequent inactivation of tumor suppressors, including CDKN2A. These data suggest that many EACs emerge not through the gradual accumulation of tumor-suppressor alterations but rather through a more direct path whereby a TP53-mutant cell undergoes genome doubling, followed by the acquisition of oncogenic amplifications.
Conflict of interest statement
Conflict of Interest Statement: The authors report no conflict of interest
The authors declare no competing financial interests.
Figures


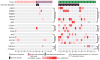

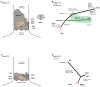


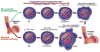
Comment in
-
Barrett oesophagus: Tracing the genetic pathway to oesophageal cancer.Nat Rev Gastroenterol Hepatol. 2015 Sep;12(9):487. doi: 10.1038/nrgastro.2015.138. Epub 2015 Aug 11. Nat Rev Gastroenterol Hepatol. 2015. PMID: 26260371 No abstract available.
References
-
- Wild CP, Hardie LJ. Reflux, Barrett’s oesophagus and adenocarcinoma: burning questions. Nat Rev Cancer. 2003;3:676–84. - PubMed
-
- Lagergren J, Bergstromeinhold R, Lingren A, Nyren O. Symptomatic Gastoroesophageal Reflux as a Risk Factor for Esophageal Adenocarcinoma. NEJM. 1999;340:825–831. - PubMed
-
- Ormsby AH, et al. The Location and Frequency of Intestinal Metaplasia at the Esophagogastric Junction in 223 Consecutive Autopsies: Implications for Patient Treatment and Preventive Strategies in Barrett’s Esophagus. Mod Pathol. 2000;6:614–620. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- K08 CA134931/CA/NCI NIH HHS/United States
- K07 CA151613/CA/NCI NIH HHS/United States
- T32 CA009676/CA/NCI NIH HHS/United States
- U54 HG0003067/HG/NHGRI NIH HHS/United States
- P50 CA127003/CA/NCI NIH HHS/United States
- T32 GM007748/GM/NIGMS NIH HHS/United States
- U54 CA163004/CA/NCI NIH HHS/United States
- L30 CA179766/CA/NCI NIH HHS/United States
- U54 HG003067/HG/NHGRI NIH HHS/United States
- T32 HL007627/HL/NHLBI NIH HHS/United States
- U54 CA163059/CA/NCI NIH HHS/United States
- P50CA127003/CA/NCI NIH HHS/United States
- P01 CA098101/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

