Using the Hepatitis C Virus RNA-Dependent RNA Polymerase as a Model to Understand Viral Polymerase Structure, Function and Dynamics
- PMID: 26193306
- PMCID: PMC4517137
- DOI: 10.3390/v7072808
Using the Hepatitis C Virus RNA-Dependent RNA Polymerase as a Model to Understand Viral Polymerase Structure, Function and Dynamics
Abstract
Viral polymerases replicate and transcribe the genomes of several viruses of global health concern such as Hepatitis C virus (HCV), human immunodeficiency virus (HIV) and Ebola virus. For this reason they are key targets for therapies to treat viral infections. Although there is little sequence similarity across the different types of viral polymerases, all of them present a right-hand shape and certain structural motifs that are highly conserved. These features allow their functional properties to be compared, with the goal of broadly applying the knowledge acquired from studying specific viral polymerases to other viral polymerases about which less is known. Here we review the structural and functional properties of the HCV RNA-dependent RNA polymerase (NS5B) in order to understand the fundamental processes underlying the replication of viral genomes. We discuss recent insights into the process by which RNA replication occurs in NS5B as well as the role that conformational changes play in this process.
Keywords: Flaviviridae; conformations; positive-strand RNA viruses.
Figures
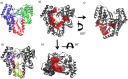
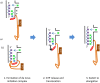
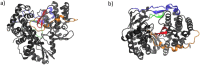
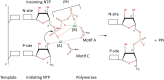
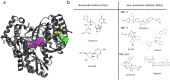
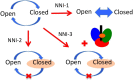
Similar articles
-
Phosphorylation of hepatitis C virus RNA polymerases ser29 and ser42 by protein kinase C-related kinase 2 regulates viral RNA replication.J Virol. 2014 Oct;88(19):11240-52. doi: 10.1128/JVI.01826-14. Epub 2014 Jul 16. J Virol. 2014. PMID: 25031343 Free PMC article.
-
A comprehensive structure-function comparison of hepatitis C virus strain JFH1 and J6 polymerases reveals a key residue stimulating replication in cell culture across genotypes.J Virol. 2011 Mar;85(6):2565-81. doi: 10.1128/JVI.02177-10. Epub 2011 Jan 5. J Virol. 2011. PMID: 21209117 Free PMC article.
-
The C-terminal transmembrane domain of hepatitis C virus (HCV) RNA polymerase is essential for HCV replication in vivo.J Virol. 2004 Apr;78(7):3797-802. doi: 10.1128/jvi.78.7.3797-3802.2004. J Virol. 2004. PMID: 15016899 Free PMC article.
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
-
Biochemical and structural analysis of the NS5B RNA-dependent RNA polymerase of the hepatitis C virus.J Viral Hepat. 2000 May;7(3):167-74. doi: 10.1046/j.1365-2893.2000.00218.x. J Viral Hepat. 2000. PMID: 10849258 Review.
Cited by
-
Hepatitis C Virus Replication.Cold Spring Harb Perspect Med. 2020 Mar 2;10(3):a037093. doi: 10.1101/cshperspect.a037093. Cold Spring Harb Perspect Med. 2020. PMID: 31570388 Free PMC article. Review.
-
Recombinant HCV NS3 and NS5B enzymes exhibit multiple posttranslational modifications for potential regulation.Virus Genes. 2019 Apr;55(2):227-232. doi: 10.1007/s11262-019-01638-2. Epub 2019 Jan 29. Virus Genes. 2019. PMID: 30694421
-
Dissecting nucleotide selectivity in viral RNA polymerases.Comput Struct Biotechnol J. 2021;19:3339-3348. doi: 10.1016/j.csbj.2021.06.005. Epub 2021 Jun 4. Comput Struct Biotechnol J. 2021. PMID: 34104356 Free PMC article. Review.
-
Inhibition of Viral RNA-Dependent RNA Polymerases by Nucleoside Inhibitors: An Illustration of the Unity and Diversity of Mechanisms.Int J Mol Sci. 2022 Oct 21;23(20):12649. doi: 10.3390/ijms232012649. Int J Mol Sci. 2022. PMID: 36293509 Free PMC article. Review.
-
Hepatitis C Viral Replication Complex.Viruses. 2021 Mar 22;13(3):520. doi: 10.3390/v13030520. Viruses. 2021. PMID: 33809897 Free PMC article. Review.
References
-
- Choi K.H. Viral Molecular Machines. Springer Science; New York, NY, USA: 2012. Viral polymerases.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

