Evolutionary determinants of cancer
- PMID: 26193902
- PMCID: PMC4539576
- DOI: 10.1158/2159-8290.CD-15-0439
Evolutionary determinants of cancer
Abstract
Our understanding of cancer is being transformed by exploring clonal diversity, drug resistance, and causation within an evolutionary framework. The therapeutic resilience of advanced cancer is a consequence of its character as a complex, dynamic, and adaptive ecosystem engendering robustness, underpinned by genetic diversity and epigenetic plasticity. The risk of mutation-driven escape by self-renewing cells is intrinsic to multicellularity but is countered by multiple restraints, facilitating increasing complexity and longevity of species. But our own species has disrupted this historical narrative by rapidly escalating intrinsic risk. Evolutionary principles illuminate these challenges and provide new avenues to explore for more effective control.
Significance: Lifetime risk of cancer now approximates to 50% in Western societies. And, despite many advances, the outcome for patients with disseminated disease remains poor, with drug resistance the norm. An evolutionary perspective may provide a clearer understanding of how cancer clones develop robustness and why, for us as a species, risk is now off the scale. And, perhaps, of what we might best do to achieve more effective control.
©2015 American Association for Cancer Research.
Figures


Revealed by single cell genetic analysis or inferred bioinformatically from in depth sequencing (9-13). The number of genetically distinct sub-clones identified depends upon the depth of genome sequencing, the number of cells interrogated and the number and nature of mutations screened. Single cell whole genome sequencing suggests that every cell is unique (16) and therefore a tumour has, in one sense, as many sub-clones as there are cells.
Variegation of genetics and clonal phylogeny architecture inferred from single cell genetics or multi-regional sequencing using maximum parsimony, other phylogenetic methods or probabilistic algorithms(192). Note that at present cancer biologists use no single or uniform platform to infer and depict phylogenetic trees. There is much to learn here from phylogenomics of species (20).
Dramatic changes in clonal structure are reflected in clonal sweeps as in metastases or drug resistant recurrence. These abrupt adaptive changes are prompted by stringent selection.
Reiterated or independent mutations in same gene in sub-clonal branches (11, 13): a result of convergent evolution and strong selective pressure favouring those mutations.
Revealed by comparative clonal phylogenetics of cancers from multiple patients with the same subtype of disease (11-13). Branching clonal architectures, reiterated mutations (or convergent evolution) in different side branches and overall uniqueness of each clonal architecture are all features of a complex adaptive or evolving biological system.
Trunkal mutations are defined as those shared by all cells in all extant sub-clones. When more than one is present, it is likely that they were acquired sequentially and some early ‘branching’ may be lost.
Xenotransplantation studies using immune-deficient mice suggest that sub-clones have variable competitive ability to generate leukaemias or cancers in vivo but in most cases this capacity resides in more than one or several sub-clones (13, 112).
Comparative genomics of matched relapse/recurrence plus diagnostic samples made it possible to detect the drug resistant mutations in the primary material (i.e. at low frequencies prior to selection) (80, 81) as well to identify minor sub-clones that have spawned metastases or relapses (22, 23, 193).
Many cancers appear to show some topographical segregation of genetically distinct sub-clones revealed in tissue sections or micro-dissected regions of primary tumours (11, 194) and in metastatic lesions (22, 23) though admixtures are also common (10).

 : at each of these levels, individual entities may aggregate into conglomerates to optimise fitness (e.g. molecular complexes, colonial bacteria and protists, social insects) in which case selection may act at the group level.
: at each of these levels, individual entities may aggregate into conglomerates to optimise fitness (e.g. molecular complexes, colonial bacteria and protists, social insects) in which case selection may act at the group level.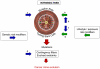
 exposures can (a) impact directly on mutation rate, e.g. genotoxic ionising radiation or genotoxic chemicals such as benzo(a)pyrene in cigarette smoke, integrated viruses, or chronic inflammation and oxidative stress. Alternatively, (b) they can indirectly increase mutation probability via replicative stress on stem cells (e.g. via toxic damage stimulated regeneration, or persistent or cyclical proliferative stimulation, e.g. hormonal stimulation or microbial infection. Excess calories (diet/exercise balance) can feed extra proliferative cycles via IGF1 levels. Inherited genetic variants
exposures can (a) impact directly on mutation rate, e.g. genotoxic ionising radiation or genotoxic chemicals such as benzo(a)pyrene in cigarette smoke, integrated viruses, or chronic inflammation and oxidative stress. Alternatively, (b) they can indirectly increase mutation probability via replicative stress on stem cells (e.g. via toxic damage stimulated regeneration, or persistent or cyclical proliferative stimulation, e.g. hormonal stimulation or microbial infection. Excess calories (diet/exercise balance) can feed extra proliferative cycles via IGF1 levels. Inherited genetic variants  can impact directly on mutation rate (c) via, for example, diminished DNA repair or can influence cancer risk ‘downstream’ by epistatic interaction with somatic mutations (d) (see text and Fig 2). Inherited variants can also impact on intrinsic risk (e,f) via their effect on exposures/lifestyle, for example via skin pigmentation (and UVB impact), nicotine addiction (and cigarette carcinogen impact) or efficacy of immune response to infections (146).
can impact directly on mutation rate (c) via, for example, diminished DNA repair or can influence cancer risk ‘downstream’ by epistatic interaction with somatic mutations (d) (see text and Fig 2). Inherited variants can also impact on intrinsic risk (e,f) via their effect on exposures/lifestyle, for example via skin pigmentation (and UVB impact), nicotine addiction (and cigarette carcinogen impact) or efficacy of immune response to infections (146).
- a
, see (75).
- b
, see references (85, 196).
- c
, see (183).
- d
, see references (13, 66, 197).
- e
, see (198).
- f
, see references (197, 199).
- g
, see (200).
- h
, see (201).
- i
, see references (40, 110)
References
-
- Stewart BW, Wild CP, editors. World Cancer Report 2014. International Agency for Research on Cancer; Lyon: 2014. - PubMed
-
- Varmus H, Kumar HS. Addressing the growing international challenge of cancer: a multinational perspective. Sci Transl Med. 2013;5(175):175cm2. - PubMed
-
- McGranahan N, Swanton C. Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution. Cancer Cell. 2015;27(1):15–26. - PubMed
-
- Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013;368:842–51. - PubMed
-
- Boveri T. The origin of malignant tumors. Baillière Tindall & Cox; London: 1929.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

