Genome-Wide Association Study of Down Syndrome-Associated Atrioventricular Septal Defects
- PMID: 26194203
- PMCID: PMC4592978
- DOI: 10.1534/g3.115.019943
Genome-Wide Association Study of Down Syndrome-Associated Atrioventricular Septal Defects
Abstract
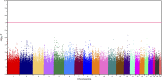
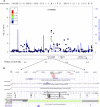
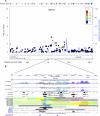
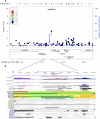
The goal of this study was to identify the contribution of common genetic variants to Down syndrome-associated atrioventricular septal defect, a severe heart abnormality. Compared with the euploid population, infants with Down syndrome, or trisomy 21, have a 2000-fold increased risk of presenting with atrioventricular septal defects. The cause of this increased risk remains elusive. Here we present data from the largest heart study conducted to date on a trisomic background by using a carefully characterized collection of individuals from extreme ends of the phenotypic spectrum. We performed a genome-wide association study using logistic regression analysis on 452 individuals with Down syndrome, consisting of 210 cases with complete atrioventricular septal defects and 242 controls with structurally normal hearts. No individual variant achieved genome-wide significance. We identified four disomic regions (1p36.3, 5p15.31, 8q22.3, and 17q22) and two trisomic regions on chromosome 21 (around PDXK and KCNJ6 genes) that merit further investigation in large replication studies. Our data show that a few common genetic variants of large effect size (odds ratio >2.0) do not account for the elevated risk of Down syndrome-associated atrioventricular septal defects. Instead, multiple variants of low-to-moderate effect sizes may contribute to this elevated risk, highlighting the complex genetic architecture of atrioventricular septal defects even in the highly susceptible Down syndrome population.
Keywords: aneuploidy; birth defect; complex trait; congenital heart defect; trisomy.
Copyright © 2015 Ramachandran et al.
Figures

References
-
- Bean L. J., Allen E. G., Tinker S. W., Hollis N. D., Locke A. E., et al. , 2011. Lack of maternal folic acid supplementation is associated with heart defects in Down syndrome: a report from the National Down Syndrome Project. Birth Defects Research (Part A). Clin. Mol. Teratol. 91: 885–893. - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
