Distinct transcriptome profiles differentiate nonsteroidal anti-inflammatory drug-dependent from nonsteroidal anti-inflammatory drug-independent food-induced anaphylaxis
- PMID: 26194548
- PMCID: PMC4715677
- DOI: 10.1016/j.jaci.2015.05.042
Distinct transcriptome profiles differentiate nonsteroidal anti-inflammatory drug-dependent from nonsteroidal anti-inflammatory drug-independent food-induced anaphylaxis
Abstract
Background: Lipid transfer protein (LTP), an abundant protein in fruits, vegetables, and nuts, is a common food allergen in Mediterranean areas causing diverse allergic reactions. Approximately 40% of food-related anaphylaxis induced by LTPs requires nonsteroidal anti-inflammatory drugs (NSAIDs) as a triggering cofactor.
Objective: We sought to better understand the determinants of NSAID-dependent and NSAID-independent LTP-induced anaphylaxis (LTP-A).
Methods: Selection of patients was based on a proved clinical history of NSAID-dependent or NSAID-independent anaphylaxis to LTPs, positive skin prick test response to LTPs, and serum LTP IgE. Whole-transcriptome (RNA sequencing) analysis of blood cells from 14 patients with NSAID-related LTP-A (NSAID-LTP-A), 7 patients with LTP-A, and 13 healthy control subjects was performed to identify distinct gene expression signatures.
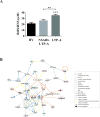
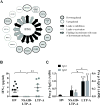
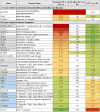
Results: Expression of genes regulating gastrointestinal epithelial renewal was altered in both patient sets, particularly in those with LTP-A, who also presented with gene expression profiles characteristic of an inflammatory syndrome. These included altered B-cell pathways, increased neutrophil activation markers, and increased reactive oxygen species levels. Increased expression of the IgG receptor (CD64) in patients with LTP-A was mirrored by the presence of LTP-specific IgG1 and IgG3. Conversely, patients with NSAID-LTP-A were characterized by reduced expression of IFN-γ-regulated genes and IFN-γ levels, as well as upregulated expression of adenosine receptor 3 (ADORA3) and genes related to adenosine metabolism.
Conclusions: Gene ontology analysis suggests disturbances in gut epithelial homeostasis in both groups with LTP-A, with potential integrity breaches in patients with LTP-A that might explain their distinct inflammatory signatures. Differential regulation in patients with LTP-A and those with NSAID-LTP-A of the IFN-γ pathway, IgG receptors, and ADORA3 might provide the pathogenic basis of their distinct responses.
Keywords: Anaphylaxis; food allergy; lipid transfer protein syndrome; nonsteroidal anti-inflammatory drugs; transcriptome analysis.
Published by Elsevier Inc.
Figures
References
-
- Sampson HA, Muñoz-Furlong A, Campbell RL, Adkinson NF, Bock SA, Branum A, et al. Second symposium on the definition and management of anaphylaxis: Summary report—Second National Institute of Allergy and Infectious Disease/Food Allergy and Anaphylaxis Network symposium. Journal of Allergy and Clinical Immunology. 2006;117:391–7. - PubMed
-
- Ben-Shoshan M, Clarke AE. Anaphylaxis: past, present and future. Allergy. 2011;66:1–14. - PubMed
-
- Cianferoni A, Novembre E, Mugnaini L, Lombardi E, Bernardini R, Pucci N, et al. Clinical features of acute anaphylaxis in patients admitted to a university hospital: an 11-year retrospective review (1985-1996) Ann Allergy Asthma Immunol. 2001;87:27–32. - PubMed
-
- Wang J, Sampson HA. Food anaphylaxis. Clin Exp Allergy. 2007;37:651–60. - PubMed
-
- Pastorello EA, Robino AM. Clinical role of lipid transfer proteins in food allergy. Mol Nutr Food Res. 2004;48:356–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

