The mutation patterns in B-cell immunoglobulin receptors reflect the influence of selection acting at multiple time-scales
- PMID: 26194756
- PMCID: PMC4528419
- DOI: 10.1098/rstb.2014.0242
The mutation patterns in B-cell immunoglobulin receptors reflect the influence of selection acting at multiple time-scales
Abstract
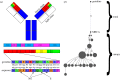
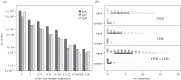
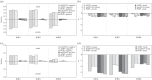
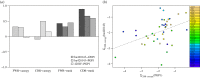
During the several-week course of an immune response, B cells undergo a process of clonal expansion, somatic hypermutation of the immunoglobulin (Ig) genes and affinity-dependent selection. Over a lifetime, each B cell may participate in multiple rounds of affinity maturation as part of different immune responses. These two time-scales for selection are apparent in the structure of B-cell lineage trees, which often contain a 'trunk' consisting of mutations that are shared across all members of a clone, and several branches that form a 'canopy' consisting of mutations that are shared by a subset of clone members. The influence of affinity maturation on the B-cell population can be inferred by analysing the pattern of somatic mutations in the Ig. While global analysis of mutation patterns has shown evidence of strong selection pressures shaping the B-cell population, the effect of different time-scales of selection and diversification has not yet been studied. Analysis of B cells from blood samples of three healthy individuals identifies a range of clone sizes with lineage trees that can contain long trunks and canopies indicating the significant diversity introduced by the affinity maturation process. We here show that observed mutation patterns in the framework regions (FWRs) are determined by an almost purely purifying selection on both short and long time-scales. By contrast, complementarity determining regions (CDRs) are affected by a combination of purifying and antigen-driven positive selection on the short term, which leads to a net positive selection in the long term. In both the FWRs and CDRs, long-term selection is strongly dependent on the heavy chain variable gene family.
Keywords: B-cell receptor; affinity maturation; antigen-driven selection; lineage trees; mutations; next generation sequencing.
© 2015 The Author(s) Published by the Royal Society. All rights reserved.
Figures

References
-
- Janeway C, Travers P, Walport M, Shlomchik M. 2004. Immunobiology, pp. 49–53, 6th edn New York, NY: Garland Science.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

