Modeling ligand recognition at the P2Y12 receptor in light of X-ray structural information
- PMID: 26194851
- PMCID: PMC4536181
- DOI: 10.1007/s10822-015-9858-z
Modeling ligand recognition at the P2Y12 receptor in light of X-ray structural information
Abstract
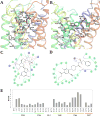
The G protein-coupled P2Y12 receptor (P2Y12R) is an important antithrombotic target and of great interest for pharmaceutical discovery. Its recently solved, highly divergent crystallographic structures in complex either with nucleotides (full or partial agonist) or with a nonnucleotide antagonist raise the question of which structure is more useful to understand ligand recognition. Therefore, we performed extensive molecular modeling studies based on these structures and mutagenesis, to predict the binding modes of major classes of P2Y12R ligands previously reported. Various nucleotide derivatives docked readily to the agonist-bound P2Y12R, but uncharged nucleotide-like antagonist ticagrelor required a hybrid receptor resembling the agonist-bound P2Y12R except for the top portion of TM6. Supervised molecular dynamics (SuMD) of ticagrelor binding indicated interactions with the extracellular regions of P2Y12R, defining possible meta-binding sites. Ureas, sulfonylureas, sulfonamides, anthraquinones and glutamic acid piperazines docked readily to the antagonist-bound P2Y12R. Docking dinucleotides at both agonist- and antagonist-bound structures suggested interactions with two P2Y12R pockets. Thus, our structure-based approach consistently rationalized the main structure-activity relationships within each ligand class, giving useful information for designing improved ligands.
Figures

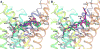
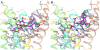

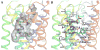






References
-
- Abbracchio MP, Burnstock G, Boeynaems JM, Barnard EA, Boyer JL, Kennedy C, Knight GE, Fumagalli M, Gachet C, Jacobson KA, Weisman GA. International Union of Pharmacology LVIII: update on the P2Y G protein-coupled nucleotide receptors: from molecular mechanisms and pathophysiology to therapy. Pharmacol Rev. 2006;58:281–341. - PMC - PubMed
-
- Burnstock G, Knight GE. Cellular distribution and functions of P2 receptor subtypes in different systems. Int Rev Cytol. 2004;240:31–304. - PubMed
-
- Hollopeter G, Jantzen HM, Vincent D, Li G, England L, Ramakrishnan V, Yang RB, Nurden P, Nurden A, Julius D, Conley PB. Identification of the platelet ADP receptor targeted by antithrombotic drugs. Nature. 2001;409:202–207. - PubMed
-
- Hourani SM, Cusack NJ. Pharmacological receptors on blood platelets. Pharmacol Rev. 1991;43:243–98. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

