IQGAP1 promotes CXCR4 chemokine receptor function and trafficking via EEA-1+ endosomes
- PMID: 26195666
- PMCID: PMC4508899
- DOI: 10.1083/jcb.201411045
IQGAP1 promotes CXCR4 chemokine receptor function and trafficking via EEA-1+ endosomes
Abstract
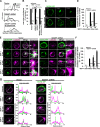
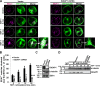
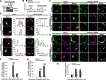
IQ motif-containing GTPase-activating protein 1 (IQGAP1) is a cytoskeleton-interacting scaffold protein. CXCR4 is a chemokine receptor that binds stromal cell-derived factor-1 (SDF-1; also known as CXCL12). Both IQGAP1 and CXCR4 are overexpressed in cancer cell types, yet it was unclear whether these molecules functionally interact. Here, we show that depleting IQGAP1 in Jurkat T leukemic cells reduced CXCR4 expression, disrupted trafficking of endocytosed CXCR4 via EEA-1(+) endosomes, and decreased efficiency of CXCR4 recycling. SDF-1-induced cell migration and activation of extracellular signal-regulated kinases 1 and 2 (ERK) MAPK were strongly inhibited, even when forced overexpression restored CXCR4 levels. Similar results were seen in KMBC and HEK293 cells. Exploring the mechanism, we found that SDF-1 treatment induced IQGAP1 binding to α-tubulin and localization to CXCR4-containing endosomes and that CXCR4-containing EEA-1(+) endosomes were abnormally located distal from the microtubule (MT)-organizing center (MTOC) in IQGAP1-deficient cells. Thus, IQGAP1 critically mediates CXCR4 cell surface expression and signaling, evidently by regulating EEA-1(+) endosome interactions with MTs during CXCR4 trafficking and recycling. IQGAP1 may similarly promote CXCR4 functions in other cancer cell types.
© 2015 Bamidele et al.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

