CHMP5 controls bone turnover rates by dampening NF-κB activity in osteoclasts
- PMID: 26195726
- PMCID: PMC4516796
- DOI: 10.1084/jem.20150407
CHMP5 controls bone turnover rates by dampening NF-κB activity in osteoclasts
Abstract
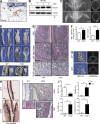
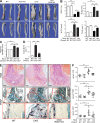
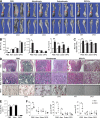
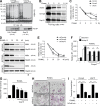
Physiological bone remodeling requires that bone formation by osteoblasts be tightly coupled to bone resorption by osteoclasts. However, relatively little is understood about how this coupling is regulated. Here, we demonstrate that modulation of NF-κB signaling in osteoclasts via a novel activity of charged multivesicular body protein 5 (CHMP5) is a key determinant of systemic rates of bone turnover. A conditional deletion of CHMP5 in osteoclasts leads to increased bone resorption by osteoclasts coupled with exuberant bone formation by osteoblasts, resembling an early onset, polyostotic form of human Paget's disease of bone (PDB). These phenotypes are reversed by haploinsufficiency for Rank, as well as by antiresorptive treatments, including alendronate, zolendronate, and OPG-Fc. Accordingly, CHMP5-deficient osteoclasts display increased RANKL-induced NF-κB activation and osteoclast differentiation. Biochemical analysis demonstrated that CHMP5 cooperates with the PDB genetic risk factor valosin-containing protein (VCP/p97) to stabilize the inhibitor of NF-κBα (IκBα), down-regulating ubiquitination of IκBα via the deubiquitinating enzyme USP15. Thus, CHMP5 tunes NF-κB signaling downstream of RANK in osteoclasts to dampen osteoclast differentiation, osteoblast coupling and bone turnover rates, and disruption of CHMP5 activity results in a PDB-like skeletal disorder.
© 2015 Greenblatt et al.
Figures





References
-
- Albagha O.M., Visconti M.R., Alonso N., Langston A.L., Cundy T., Dargie R., Dunlop M.G., Fraser W.D., Hooper M.J., Isaia G., et al. . 2010. Genome-wide association study identifies variants at CSF1, OPTN and TNFRSF11A as genetic risk factors for Paget’s disease of bone. Nat. Genet. 42:520–524. 10.1038/ng.562 - DOI - PMC - PubMed
-
- Albagha O.M., Wani S.E., Visconti M.R., Alonso N., Goodman K., Brandi M.L., Cundy T., Chung P.Y., Dargie R., Devogelaer J.P., et al. . Genetic Determinants of Paget’s Disease (GDPD) Consortium. 2011. Genome-wide association identifies three new susceptibility loci for Paget’s disease of bone. Nat. Genet. 43:685–689. 10.1038/ng.845 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

