Entner-Doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1
- PMID: 26195800
- PMCID: PMC4534283
- DOI: 10.1073/pnas.1507049112
Entner-Doudoroff pathway for sulfoquinovose degradation in Pseudomonas putida SQ1
Abstract
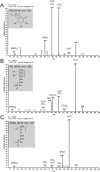
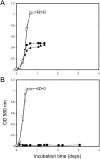
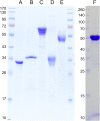
Sulfoquinovose (SQ; 6-deoxy-6-sulfoglucose) is the polar head group of the plant sulfolipid SQ-diacylglycerol, and SQ comprises a major proportion of the organosulfur in nature, where it is degraded by bacteria. A first degradation pathway for SQ has been demonstrated recently, a "sulfoglycolytic" pathway, in addition to the classical glycolytic (Embden-Meyerhof) pathway in Escherichia coli K-12; half of the carbon of SQ is abstracted as dihydroxyacetonephosphate (DHAP) and used for growth, whereas a C3-organosulfonate, 2,3-dihydroxypropane sulfonate (DHPS), is excreted. The environmental isolate Pseudomonas putida SQ1 is also able to use SQ for growth, and excretes a different C3-organosulfonate, 3-sulfolactate (SL). In this study, we revealed the catabolic pathway for SQ in P. putida SQ1 through differential proteomics and transcriptional analyses, by in vitro reconstitution of the complete pathway by five heterologously produced enzymes, and by identification of all four organosulfonate intermediates. The pathway follows a reaction sequence analogous to the Entner-Doudoroff pathway for glucose-6-phosphate: It involves an NAD(+)-dependent SQ dehydrogenase, 6-deoxy-6-sulfogluconolactone (SGL) lactonase, 6-deoxy-6-sulfogluconate (SG) dehydratase, and 2-keto-3,6-dideoxy-6-sulfogluconate (KDSG) aldolase. The aldolase reaction yields pyruvate, which supports growth of P. putida, and 3-sulfolactaldehyde (SLA), which is oxidized to SL by an NAD(P)(+)-dependent SLA dehydrogenase. All five enzymes are encoded in a single gene cluster that includes, for example, genes for transport and regulation. Homologous gene clusters were found in genomes of other P. putida strains, in other gamma-Proteobacteria, and in beta- and alpha-Proteobacteria, for example, in genomes of Enterobacteria, Vibrio, and Halomonas species, and in typical soil bacteria, such as Burkholderia, Herbaspirillum, and Rhizobium.
Keywords: 6-deoxy-6-sulfoglucose; bacterial biodegradation; organosulfonate; sulfolipid; sulfur cycle.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Benson AA. The plant sulfolipid. Adv Lipid Res. 1963;1:387–394. - PubMed
-
- Harwood JL, Nicholls RG. The plant sulpholipid—A major component of the sulphur cycle. Biochem Soc Trans. 1979;7(2):440–447. - PubMed
-
- Benning C. Biosynthesis and function of the sulfolipid sulfoquinovosyl diacylglycerol. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:53–75. - PubMed
-
- Martelli HL, Benson AA. Sulfocarbohydrate metabolism. I. Bacterial production and utilization of sulfoacetate. Biochim Biophys Acta. 1964;93(1):169–171. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

