Nested PCR Biases in Interpreting Microbial Community Structure in 16S rRNA Gene Sequence Datasets
- PMID: 26196512
- PMCID: PMC4509648
- DOI: 10.1371/journal.pone.0132253
Nested PCR Biases in Interpreting Microbial Community Structure in 16S rRNA Gene Sequence Datasets
Abstract
Background: Sequencing of the PCR-amplified 16S rRNA gene has become a common approach to microbial community investigations in the fields of human health and environmental sciences. This approach, however, is difficult when the amount of DNA is too low to be amplified by standard PCR. Nested PCR can be employed as it can amplify samples with DNA concentration several-fold lower than standard PCR. However, potential biases with nested PCRs that could affect measurement of community structure have received little attention.
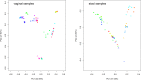
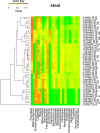
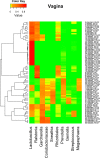
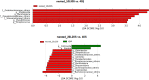
Results: In this study, we used 17 DNAs extracted from vaginal swabs and 12 DNAs extracted from stool samples to study the influence of nested PCR amplification of the 16S rRNA gene on the estimation of microbial community structure using Illumina MiSeq sequencing. Nested and standard PCR methods were compared on alpha- and beta-diversity metrics and relative abundances of bacterial genera. The effects of number of cycles in the first round of PCR (10 vs. 20) and microbial diversity (relatively low in vagina vs. high in stool) were also investigated. Vaginal swab samples showed no significant difference in alpha diversity or community structure between nested PCR and standard PCR (one round of 40 cycles). Stool samples showed significant differences in alpha diversity (except Shannon's index) and relative abundance of 13 genera between nested PCR with 20 cycles in the first round and standard PCR (P<0.01), but not between nested PCR with 10 cycles in the first round and standard PCR. Operational taxonomic units (OTUs) that had low relative abundance (sum of relative abundance <0.167) accounted for most of the distortion (>27% of total OTUs in stool).
Conclusions: Nested PCR introduced bias in estimated diversity and community structure. The bias was more significant for communities with relatively higher diversity and when more cycles were applied in the first round of PCR. We conclude that nested PCR could be used when standard PCR does not work. However, rare taxa detected by nested PCR should be validated by other technologies.
Conflict of interest statement
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

