Genetic evidence for two founding populations of the Americas
- PMID: 26196601
- PMCID: PMC4982469
- DOI: 10.1038/nature14895
Genetic evidence for two founding populations of the Americas
Abstract
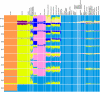
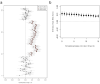
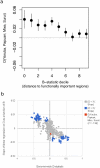
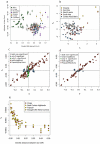
Genetic studies have consistently indicated a single common origin of Native American groups from Central and South America. However, some morphological studies have suggested a more complex picture, whereby the northeast Asian affinities of present-day Native Americans contrast with a distinctive morphology seen in some of the earliest American skeletons, which share traits with present-day Australasians (indigenous groups in Australia, Melanesia, and island Southeast Asia). Here we analyse genome-wide data to show that some Amazonian Native Americans descend partly from a Native American founding population that carried ancestry more closely related to indigenous Australians, New Guineans and Andaman Islanders than to any present-day Eurasians or Native Americans. This signature is not present to the same extent, or at all, in present-day Northern and Central Americans or in a ∼12,600-year-old Clovis-associated genome, suggesting a more diverse set of founding populations of the Americas than previously accepted.
Figures

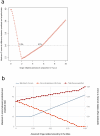

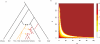
References
-
- Neves W, Pucciarelli H. American Journal of Physical Anthropology. WILEY-LISS DIV JOHN WILEY & SONS INC; 605 THIRD AVE, NEW YORK, NY 10158-0012: pp. 274–274.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

