The Orphan Nuclear Receptor TLX Is an Enhancer of STAT1-Mediated Transcription and Immunity to Toxoplasma gondii
- PMID: 26196739
- PMCID: PMC4509904
- DOI: 10.1371/journal.pbio.1002200
The Orphan Nuclear Receptor TLX Is an Enhancer of STAT1-Mediated Transcription and Immunity to Toxoplasma gondii
Abstract
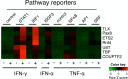
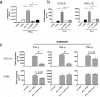
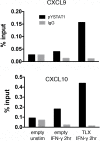
The protozoan parasite, Toxoplasma, like many intracellular pathogens, suppresses interferon gamma (IFN-γ)-induced signal transducer and activator of transcription 1 (STAT1) activity. We exploited this well-defined host-pathogen interaction as the basis for a high-throughput screen, identifying nine transcription factors that enhance STAT1 function in the nucleus, including the orphan nuclear hormone receptor TLX. Expression profiling revealed that upon IFN-γ treatment TLX enhances the output of a subset of IFN-γ target genes, which we found is dependent on TLX binding at those loci. Moreover, infection of TLX deficient mice with the intracellular parasite Toxoplasma results in impaired production of the STAT1-dependent cytokine interleukin-12 by dendritic cells and increased parasite burden in the brain during chronic infection. These results demonstrate a previously unrecognized role for this orphan nuclear hormone receptor in regulating STAT1 signaling and host defense and reveal that STAT1 activity can be modulated in a context-specific manner by such "modifiers."
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Comment in
-
Tuning up STAT1.PLoS Biol. 2015 Jul 21;13(7):e1002201. doi: 10.1371/journal.pbio.1002201. eCollection 2015 Jul. PLoS Biol. 2015. PMID: 26196960 Free PMC article.
References
-
- Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, et al. (1993) Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science 259: 1739–1742. - PubMed
-
- Durbin JE, Hackenmiller R, Simon MC, Levy DE (1996) Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84: 443–450. - PubMed
-
- Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, et al. (1996) Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84: 431–442. - PubMed
-
- Newport MJ, Huxley CM, Huston S, Hawrylowicz CM, Oostra BA, et al. (1996) A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med 335: 1941–1949. - PubMed
-
- Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, et al. (2003) Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet 33: 388–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

