K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
- PMID: 26197391
- PMCID: PMC4510548
- DOI: 10.1371/journal.ppat.1005051
K-bZIP Mediated SUMO-2/3 Specific Modification on the KSHV Genome Negatively Regulates Lytic Gene Expression and Viral Reactivation
Abstract
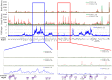
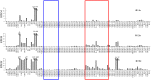
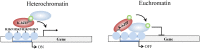
SUMOylation is associated with epigenetic regulation of chromatin structure and transcription. Epigenetic modifications of herpesviral genomes accompany the transcriptional switch of latent and lytic genes during the virus life cycle. Here, we report a genome-wide comparison of SUMO paralog modification on the KSHV genome. Using chromatin immunoprecipitation in conjunction with high-throughput sequencing, our study revealed highly distinct landscape changes of SUMO paralog genomic modifications associated with KSHV reactivation. A rapid and widespread deposition of SUMO-2/3, compared with SUMO-1, modification across the KSHV genome upon reactivation was observed. Interestingly, SUMO-2/3 enrichment was inversely correlated with H3K9me3 mark after reactivation, indicating that SUMO-2/3 may be responsible for regulating the expression of viral genes located in low heterochromatin regions during viral reactivation. RNA-sequencing analysis showed that the SUMO-2/3 enrichment pattern positively correlated with KSHV gene expression profiles. Activation of KSHV lytic genes located in regions with high SUMO-2/3 enrichment was enhanced by SUMO-2/3 knockdown. These findings suggest that SUMO-2/3 viral chromatin modification contributes to the diminution of viral gene expression during reactivation. Our previous study identified a SUMO-2/3-specific viral E3 ligase, K-bZIP, suggesting a potential role of this enzyme in regulating SUMO-2/3 enrichment and viral gene repression. Consistent with this prediction, higher K-bZIP binding on SUMO-2/3 enrichment region during reactivation was observed. Moreover, a K-bZIP SUMO E3 ligase dead mutant, K-bZIP-L75A, in the viral context, showed no SUMO-2/3 enrichment on viral chromatin and higher expression of viral genes located in SUMO-2/3 enriched regions during reactivation. Importantly, virus production significantly increased in both SUMO-2/3 knockdown and KSHV K-bZIP-L75A mutant cells. These results indicate that SUMO-2/3 modification of viral chromatin may function to counteract KSHV reactivation. As induction of herpesvirus reactivation may activate cellular antiviral regimes, our results suggest that development of viral SUMO E3 ligase specific inhibitors may be an avenue for anti-virus therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

