Poised chromatin and bivalent domains facilitate the mitosis-to-meiosis transition in the male germline
- PMID: 26198001
- PMCID: PMC4508805
- DOI: 10.1186/s12915-015-0159-8
Poised chromatin and bivalent domains facilitate the mitosis-to-meiosis transition in the male germline
Abstract
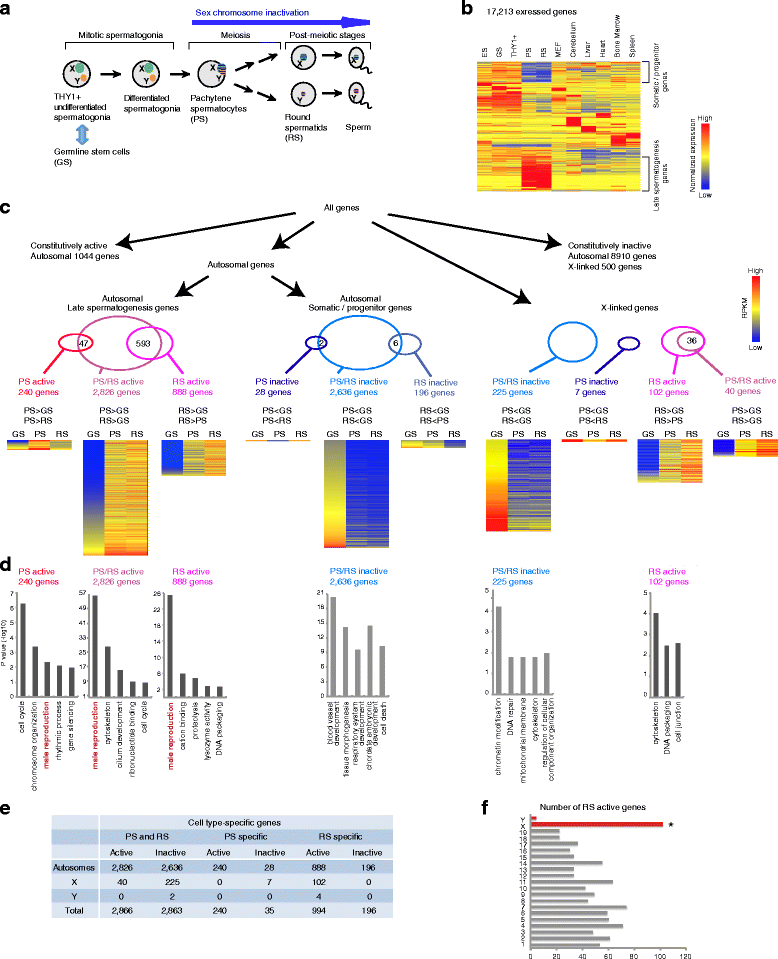
Background: The male germline transcriptome changes dramatically during the mitosis-to-meiosis transition to activate late spermatogenesis genes and to transiently suppress genes commonly expressed in somatic lineages and spermatogenesis progenitor cells, termed somatic/progenitor genes.
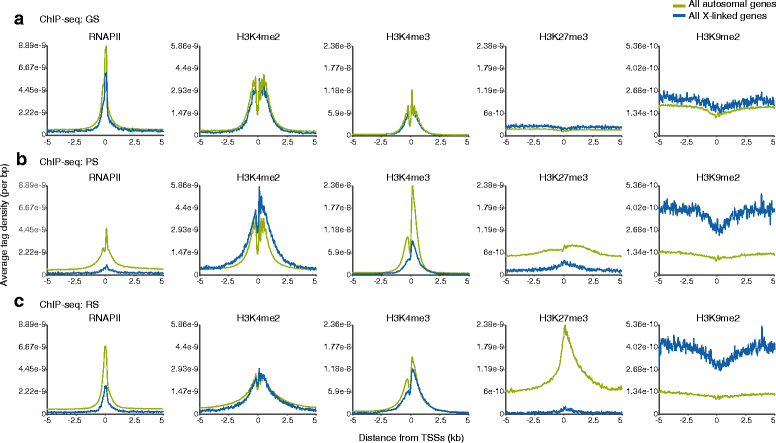
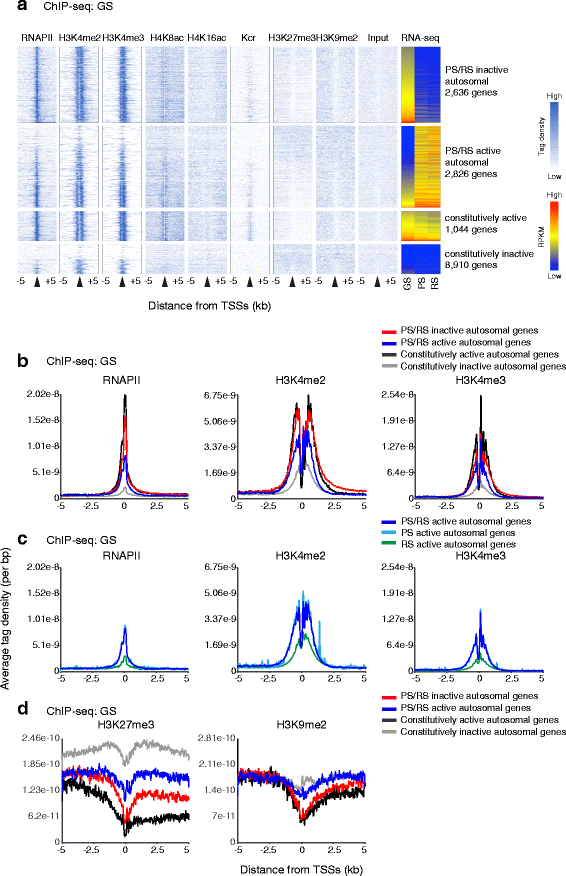
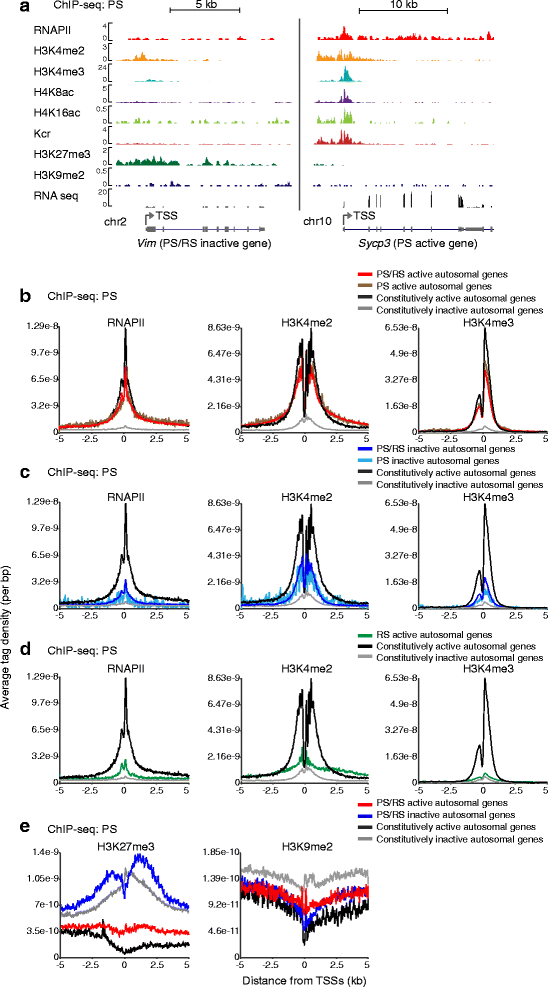
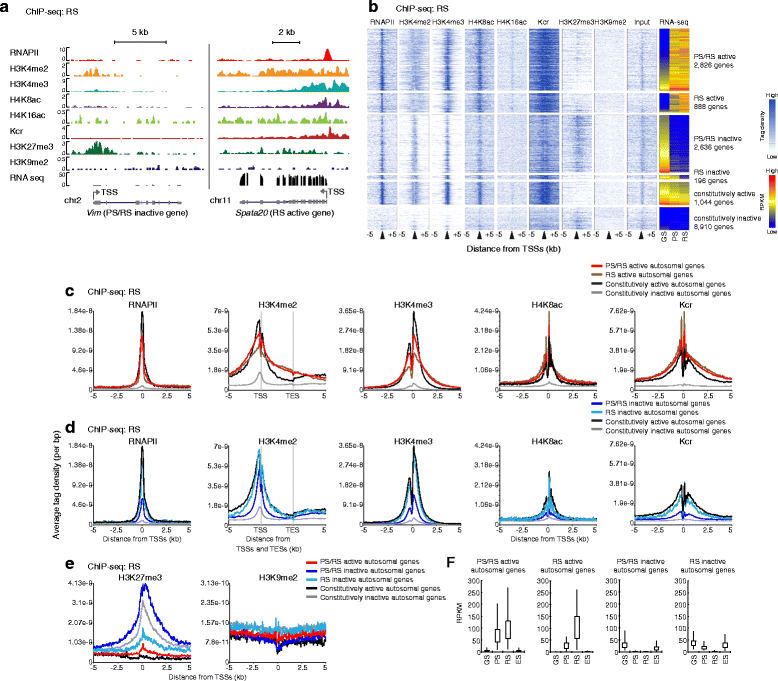
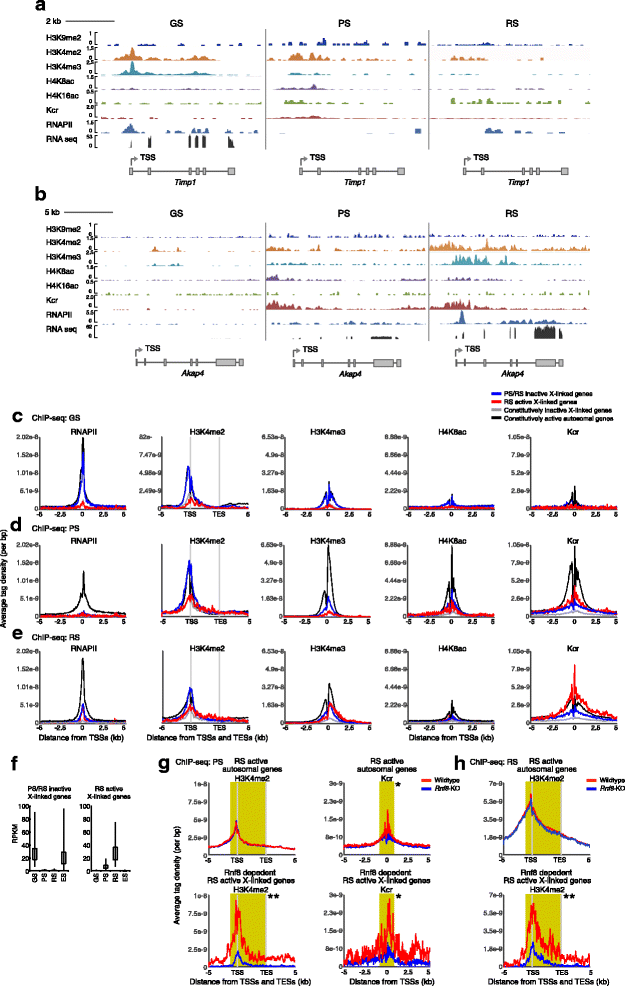
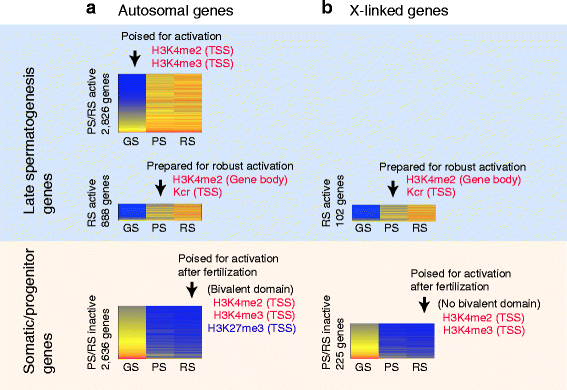
Results: These changes reflect epigenetic regulation. Induction of late spermatogenesis genes during spermatogenesis is facilitated by poised chromatin established in the stem cell phases of spermatogonia, whereas silencing of somatic/progenitor genes during meiosis and postmeiosis is associated with formation of bivalent domains which also allows the recovery of the somatic/progenitor program after fertilization. Importantly, during spermatogenesis mechanisms of epigenetic regulation on sex chromosomes are different from autosomes: X-linked somatic/progenitor genes are suppressed by meiotic sex chromosome inactivation without deposition of H3K27me3.
Conclusions: Our results suggest that bivalent H3K27me3 and H3K4me2/3 domains are not limited to developmental promoters (which maintain bivalent domains that are silent throughout the reproductive cycle), but also underlie reversible silencing of somatic/progenitor genes during the mitosis-to-meiosis transition in late spermatogenesis.
Figures
Similar articles
-
Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis.Nucleic Acids Res. 2018 Jan 25;46(2):593-608. doi: 10.1093/nar/gkx1052. Nucleic Acids Res. 2018. PMID: 29126117 Free PMC article.
-
Expression and epigenomic landscape of the sex chromosomes in mouse post-meiotic male germ cells.Epigenetics Chromatin. 2016 Oct 27;9:47. doi: 10.1186/s13072-016-0099-8. eCollection 2016. Epigenetics Chromatin. 2016. PMID: 27795737 Free PMC article.
-
Polycomb protein SCML2 facilitates H3K27me3 to establish bivalent domains in the male germline.Proc Natl Acad Sci U S A. 2018 May 8;115(19):4957-4962. doi: 10.1073/pnas.1804512115. Epub 2018 Apr 23. Proc Natl Acad Sci U S A. 2018. PMID: 29686098 Free PMC article.
-
Histone crotonylation specifically marks the haploid male germ cell gene expression program: post-meiotic male-specific gene expression.Bioessays. 2012 Mar;34(3):187-93. doi: 10.1002/bies.201100141. Epub 2011 Dec 15. Bioessays. 2012. PMID: 22170506 Review.
-
Epigenetic mechanisms of gene regulation during mammalian spermatogenesis.Epigenetics. 2008 Jan-Feb;3(1):21-8. doi: 10.4161/epi.3.1.5555. Epigenetics. 2008. PMID: 18416029 Review.
Cited by
-
Transcriptomics of Meiosis in the Male Mouse.Front Cell Dev Biol. 2021 Mar 5;9:626020. doi: 10.3389/fcell.2021.626020. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33748111 Free PMC article. Review.
-
The conserved RNA helicase YTHDC2 regulates the transition from proliferation to differentiation in the germline.Elife. 2017 Oct 31;6:e26116. doi: 10.7554/eLife.26116. Elife. 2017. PMID: 29087293 Free PMC article.
-
Polycomb protein SCML2 mediates paternal epigenetic inheritance through sperm chromatin.Nucleic Acids Res. 2023 Jul 21;51(13):6668-6683. doi: 10.1093/nar/gkad479. Nucleic Acids Res. 2023. PMID: 37283086 Free PMC article.
-
Quantitative CUT&Tag for Epigenomic Profiling of Mouse Germ Cells.Methods Mol Biol. 2025;2954:27-47. doi: 10.1007/978-1-0716-4698-4_2. Methods Mol Biol. 2025. PMID: 40601268
-
Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis.Nucleic Acids Res. 2018 Jan 25;46(2):593-608. doi: 10.1093/nar/gkx1052. Nucleic Acids Res. 2018. PMID: 29126117 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

