STAT3 regulated ARF expression suppresses prostate cancer metastasis
- PMID: 26198641
- PMCID: PMC4525303
- DOI: 10.1038/ncomms8736
STAT3 regulated ARF expression suppresses prostate cancer metastasis
Erratum in
-
Erratum: STAT3 regulated ARF expression suppresses prostate cancer metastasis.Nat Commun. 2015 Oct 26;6:8802. doi: 10.1038/ncomms9802. Nat Commun. 2015. PMID: 26499136 Free PMC article. No abstract available.
Abstract
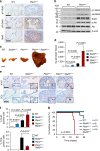
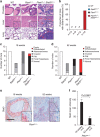
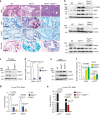
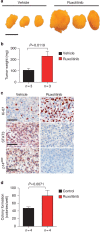
Prostate cancer (PCa) is the most prevalent cancer in men. Hyperactive STAT3 is thought to be oncogenic in PCa. However, targeting of the IL-6/STAT3 axis in PCa patients has failed to provide therapeutic benefit. Here we show that genetic inactivation of Stat3 or IL-6 signalling in a Pten-deficient PCa mouse model accelerates cancer progression leading to metastasis. Mechanistically, we identify p19(ARF) as a direct Stat3 target. Loss of Stat3 signalling disrupts the ARF-Mdm2-p53 tumour suppressor axis bypassing senescence. Strikingly, we also identify STAT3 and CDKN2A mutations in primary human PCa. STAT3 and CDKN2A deletions co-occurred with high frequency in PCa metastases. In accordance, loss of STAT3 and p14(ARF) expression in patient tumours correlates with increased risk of disease recurrence and metastatic PCa. Thus, STAT3 and ARF may be prognostic markers to stratify high from low risk PCa patients. Our findings challenge the current discussion on therapeutic benefit or risk of IL-6/STAT3 inhibition.
Figures



Comment in
-
Prostate cancer: On the right path--Stat3 signalling controls the ARF-Mdm2-p53 tumour-suppressor pathway.Nat Rev Urol. 2015 Oct;12(10):535. doi: 10.1038/nrurol.2015.205. Epub 2015 Aug 11. Nat Rev Urol. 2015. PMID: 26260883 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

