Computational redesign of the lipid-facing surface of the outer membrane protein OmpA
- PMID: 26199411
- PMCID: PMC4534290
- DOI: 10.1073/pnas.1501836112
Computational redesign of the lipid-facing surface of the outer membrane protein OmpA
Abstract
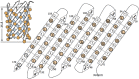
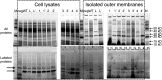
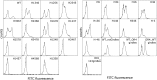
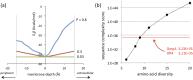
Advances in computational design methods have made possible extensive engineering of soluble proteins, but designed β-barrel membrane proteins await improvements in our understanding of the sequence determinants of folding and stability. A subset of the amino acid residues of membrane proteins interact with the cell membrane, and the design rules that govern this lipid-facing surface are poorly understood. We applied a residue-level depth potential for β-barrel membrane proteins to the complete redesign of the lipid-facing surface of Escherichia coli OmpA. Initial designs failed to fold correctly, but reversion of a small number of mutations indicated by backcross experiments yielded designs with substitutions to up to 60% of the surface that did support folding and membrane insertion.
Keywords: OmpA; membrane proteins; protein design; statistical potential; β-barrel.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Folding intermediates of a beta-barrel membrane protein. Kinetic evidence for a multi-step membrane insertion mechanism.Biochemistry. 1996 Oct 8;35(40):12993-3000. doi: 10.1021/bi961478b. Biochemistry. 1996. PMID: 8855933
-
Association of spin-labeled lipids with beta-barrel proteins from the outer membrane of Escherichia coli.Biochemistry. 2004 Sep 21;43(37):11630-6. doi: 10.1021/bi048858e. Biochemistry. 2004. PMID: 15362847
-
Concatemers of Outer Membrane Protein A Take Detours in the Folding Landscape.Biochemistry. 2016 Dec 27;55(51):7123-7140. doi: 10.1021/acs.biochem.6b01153. Epub 2016 Dec 14. Biochemistry. 2016. PMID: 27973779
-
Folding kinetics of the outer membrane proteins OmpA and FomA into phospholipid bilayers.Chem Phys Lipids. 2006 Jun;141(1-2):30-47. doi: 10.1016/j.chemphyslip.2006.02.004. Epub 2006 Mar 20. Chem Phys Lipids. 2006. PMID: 16581049 Review.
-
Augmenting β-augmentation: structural basis of how BamB binds BamA and may support folding of outer membrane proteins.J Mol Biol. 2011 Mar 11;406(5):659-66. doi: 10.1016/j.jmb.2011.01.002. Epub 2011 Jan 12. J Mol Biol. 2011. PMID: 21236263 Review.
Cited by
-
Evolutionary Engineering a Larger Porin Using a Loop-to-Hairpin Mechanism.J Mol Biol. 2023 Nov 15;435(22):168292. doi: 10.1016/j.jmb.2023.168292. Epub 2023 Sep 26. J Mol Biol. 2023. PMID: 37769963 Free PMC article.
-
Designing minimalist membrane proteins.Biochem Soc Trans. 2019 Oct 31;47(5):1233-1245. doi: 10.1042/BST20190170. Biochem Soc Trans. 2019. PMID: 31671181 Free PMC article. Review.
-
ProteinMPNN Recovers Complex Sequence Properties of Transmembrane β-barrels.bioRxiv [Preprint]. 2024 Feb 1:2024.01.16.575764. doi: 10.1101/2024.01.16.575764. bioRxiv. 2024. PMID: 38352434 Free PMC article. Preprint.
-
De novo design of transmembrane β barrels.Science. 2021 Feb 19;371(6531):eabc8182. doi: 10.1126/science.abc8182. Science. 2021. PMID: 33602829 Free PMC article.
-
Dawn of a New Era for Membrane Protein Design.Biodes Res. 2022 Apr 15;2022:9791435. doi: 10.34133/2022/9791435. eCollection 2022. Biodes Res. 2022. PMID: 37850134 Free PMC article. Review.
References
-
- Wimley WC. The versatile β-barrel membrane protein. Curr Opin Struct Biol. 2003;13(4):404–411. - PubMed
-
- Daugherty PS, Olsen MJ, Iverson BL, Georgiou G. Development of an optimized expression system for the screening of antibody libraries displayed on the Escherichia coli surface. Protein Eng. 1999;12(7):613–621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

