Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis
- PMID: 26199412
- PMCID: PMC4534209
- DOI: 10.1073/pnas.1510510112
Simultaneous deletion of the methylcytosine oxidases Tet1 and Tet3 increases transcriptome variability in early embryogenesis
Abstract
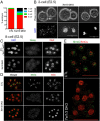
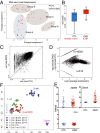
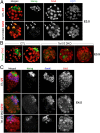
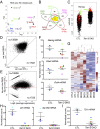
Dioxygenases of the TET (Ten-Eleven Translocation) family produce oxidized methylcytosines, intermediates in DNA demethylation, as well as new epigenetic marks. Here we show data suggesting that TET proteins maintain the consistency of gene transcription. Embryos lacking Tet1 and Tet3 (Tet1/3 DKO) displayed a strong loss of 5-hydroxymethylcytosine (5hmC) and a concurrent increase in 5-methylcytosine (5mC) at the eight-cell stage. Single cells from eight-cell embryos and individual embryonic day 3.5 blastocysts showed unexpectedly variable gene expression compared with controls, and this variability correlated in blastocysts with variably increased 5mC/5hmC in gene bodies and repetitive elements. Despite the variability, genes encoding regulators of cholesterol biosynthesis were reproducibly down-regulated in Tet1/3 DKO blastocysts, resulting in a characteristic phenotype of holoprosencephaly in the few embryos that survived to later stages. Thus, TET enzymes and DNA cytosine modifications could directly or indirectly modulate transcriptional noise, resulting in the selective susceptibility of certain intracellular pathways to regulation by TET proteins.
Keywords: 5-hydroxymethylcytosine; 5hmC; DNA methylation; TET methylcytosine oxidases; cholesterol biosynthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Comparative dynamics of 5-methylcytosine reprogramming and TET family expression during preimplantation mammalian development in mouse and sheep.Theriogenology. 2017 Feb;89:86-96. doi: 10.1016/j.theriogenology.2016.10.010. Epub 2016 Oct 20. Theriogenology. 2017. PMID: 28043375
-
Ascorbic acid improves parthenogenetic embryo development through TET proteins in mice.Biosci Rep. 2019 Jan 11;39(1):BSR20181730. doi: 10.1042/BSR20181730. Print 2019 Jan 31. Biosci Rep. 2019. PMID: 30567727 Free PMC article.
-
Combined deficiency of Tet1 and Tet2 causes epigenetic abnormalities but is compatible with postnatal development.Dev Cell. 2013 Feb 11;24(3):310-23. doi: 10.1016/j.devcel.2012.12.015. Epub 2013 Jan 24. Dev Cell. 2013. PMID: 23352810 Free PMC article.
-
Tet family of 5-methylcytosine dioxygenases in mammalian development.J Hum Genet. 2013 Jul;58(7):421-7. doi: 10.1038/jhg.2013.63. Epub 2013 May 30. J Hum Genet. 2013. PMID: 23719188 Free PMC article. Review.
-
Role of ten-eleven translocation proteins and 5-hydroxymethylcytosine in hepatocellular carcinoma.Cell Prolif. 2019 Jul;52(4):e12626. doi: 10.1111/cpr.12626. Epub 2019 Apr 29. Cell Prolif. 2019. PMID: 31033072 Free PMC article. Review.
Cited by
-
Repression of FGF signaling is responsible for Dnmt3b inhibition and impaired de novo DNA methylation during early development of in vitro fertilized embryos.Int J Biol Sci. 2020 Oct 3;16(15):3085-3099. doi: 10.7150/ijbs.51607. eCollection 2020. Int J Biol Sci. 2020. PMID: 33061820 Free PMC article.
-
TET1 regulates gene expression and repression of endogenous retroviruses independent of DNA demethylation.Nucleic Acids Res. 2022 Aug 26;50(15):8491-8511. doi: 10.1093/nar/gkac642. Nucleic Acids Res. 2022. PMID: 35904814 Free PMC article.
-
TET deficiency perturbs mature B cell homeostasis and promotes oncogenesis associated with accumulation of G-quadruplex and R-loop structures.Nat Immunol. 2022 Jan;23(1):99-108. doi: 10.1038/s41590-021-01087-w. Epub 2021 Dec 22. Nat Immunol. 2022. PMID: 34937926 Free PMC article.
-
Control of Foxp3 stability through modulation of TET activity.J Exp Med. 2016 Mar 7;213(3):377-97. doi: 10.1084/jem.20151438. Epub 2016 Feb 22. J Exp Med. 2016. PMID: 26903244 Free PMC article.
-
Advance in the Role of Epigenetic Reprogramming in Somatic Cell Nuclear Transfer-Mediated Embryonic Development.Stem Cells Int. 2021 Feb 4;2021:6681337. doi: 10.1155/2021/6681337. eCollection 2021. Stem Cells Int. 2021. PMID: 33628270 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

