O6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity
- PMID: 26199454
- PMCID: PMC4536324
- DOI: 10.1261/rna.052464.115
O6-Methylguanosine leads to position-dependent effects on ribosome speed and fidelity
Abstract
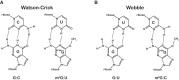
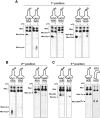
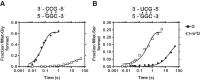
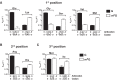
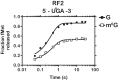
Nucleic acids are under constant assault from endogenous and environmental agents that alter their physical and chemical properties. O6-methylation of guanosine (m(6)G) is particularly notable for its high mutagenicity, pairing with T, during DNA replication. Yet, while m(6)G accumulates in both DNA and RNA, little is known about its effects on RNA. Here, we investigate the effects of m(6)G on the decoding process, using a reconstituted bacterial translation system. m(6)G at the first and third position of the codon decreases the accuracy of tRNA selection. The ribosome readily incorporates near-cognate aminoacyl-tRNAs (aa-tRNAs) by forming m(6)G-uridine codon-anticodon pairs. Surprisingly, the introduction of m(6)G to the second position of the codon does not promote miscoding, but instead slows the observed rates of peptide-bond formation by >1000-fold for cognate aa-tRNAs without altering the rates for near-cognate aa-tRNAs. These in vitro observations were recapitulated in eukaryotic extracts and HEK293 cells. Interestingly, the analogous modification N6-methyladenosine (m(6)A) at the second position has only a minimal effect on tRNA selection, suggesting that the effects on tRNA selection seen with m(6)G are due to altered geometry of the base pair. Given that the m6G:U base pair is predicted to be nearly indistinguishable from a Watson-Crick base pair, our data suggest that the decoding center of the ribosome is extremely sensitive to changes at the second position. Our data, apart from highlighting the deleterious effects that these adducts pose to cellular fitness, shed new insight into decoding and the process by which the ribosome recognizes codon-anticodon pairs.
Keywords: O6-methylguanosine; RNA damage; decoding; ribosome; translation.
© 2015 Hudson and Zaher; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures



References
-
- Aas PA, Otterlei M, Falnes PO, Vågbø CB, Skorpen F, Akbari M, Sundheim O, Bjørås M, Slupphaug G, Seeberg E, et al. 2003. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421: 859–863. - PubMed
-
- Carter AP, Clemons WM Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501. - PubMed
-
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. 2012. A new understanding of the decoding principle on the ribosome. Nature 484: 256–259. - PubMed
-
- Demple B, Jacobsson A, Olsson M, Robins P, Lindahl T. 1982. Repair of alkylated DNA in Escherichia coli. Physical properties of O6-methylguanine-DNA methyltransferase. J Biol Chem 257: 13776–13780. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
