Targeting the heparin-binding domain of fibroblast growth factor receptor 1 as a potential cancer therapy
- PMID: 26201468
- PMCID: PMC4511971
- DOI: 10.1186/s12943-015-0391-4
Targeting the heparin-binding domain of fibroblast growth factor receptor 1 as a potential cancer therapy
Abstract
Background: Aberrant activation of fibroblast growth factor receptors (FGFRs) deregulates cell proliferation and promotes cell survival, and may predispose to tumorigenesis. Therefore, selective inactivation of FGFRs is an important strategy for cancer therapy. Here as a proof-of-concept study, we developed a FGFR1 neutralizing antisera, IMB-R1, employing a novel strategy aimed at preventing the access of essential heparan sulfate (HS) co-receptors to the heparin-binding domain on FGFR1.
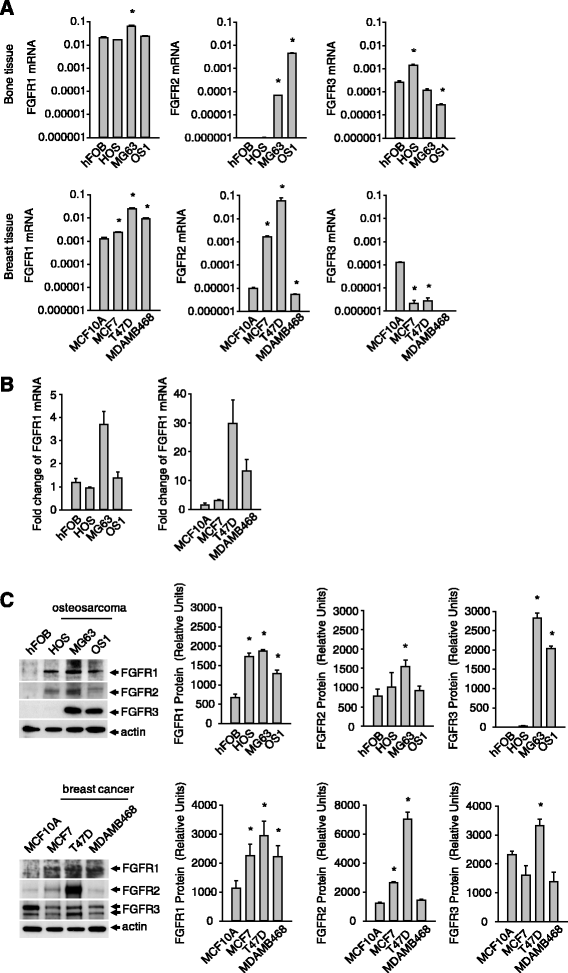
Methods: The mRNA and protein expression level of FGFR1 and other FGFRs were examined in several lines of breast cancer and osteosarcoma cells and corresponding normal cells using Taqman real-time quantitative PCR and Western blot analysis. The specificity of IMB-R1 against FGFR1 was assessed with various ELISA-based approaches and Receptor Tyrosine Kinase array. Proliferation assay and apoptosis analysis were performed to assess the effect of IMB-R1 on cancer cell growth and apoptosis, respectively, in comparison with known FGFR1 inhibitors. The IMB-R1 induced alteration of intracellular signaling and gene expression were analysed using Western blot and microarray approaches. Immunohistochemical staining of FGFR1 using IMB-R1 were carried out in different cancer tissues from clinical patients. Throughout the study, statistical differences were determined by Student's t test where appropriate and reported when a p value was less than 0.05.
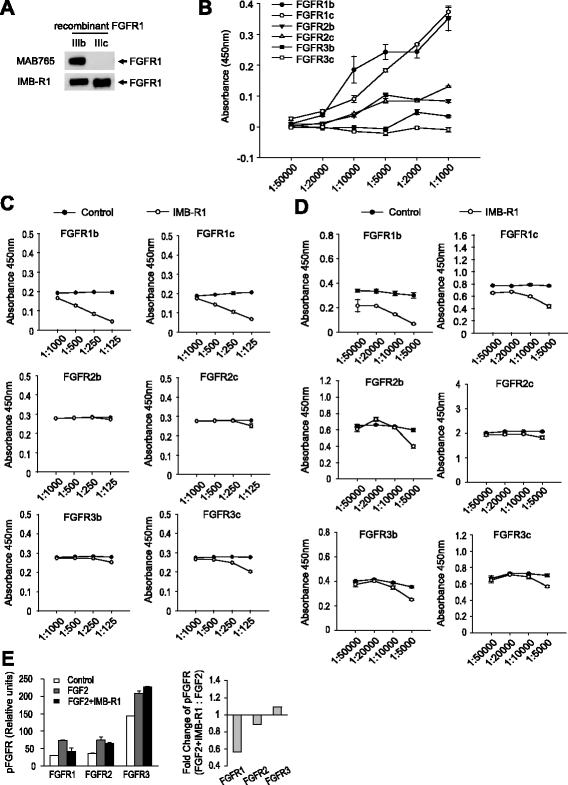
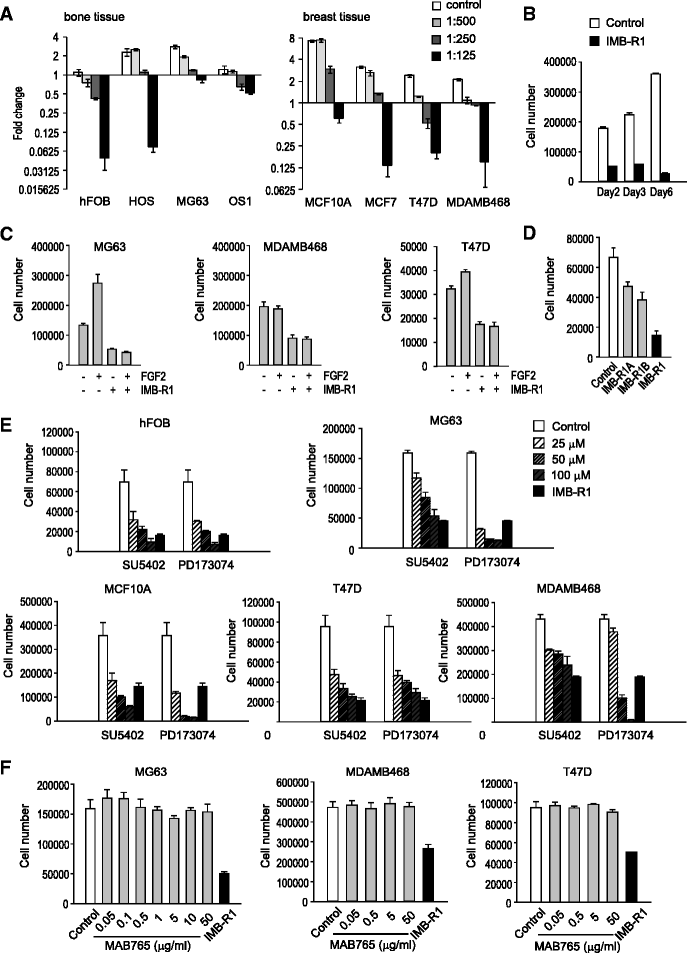
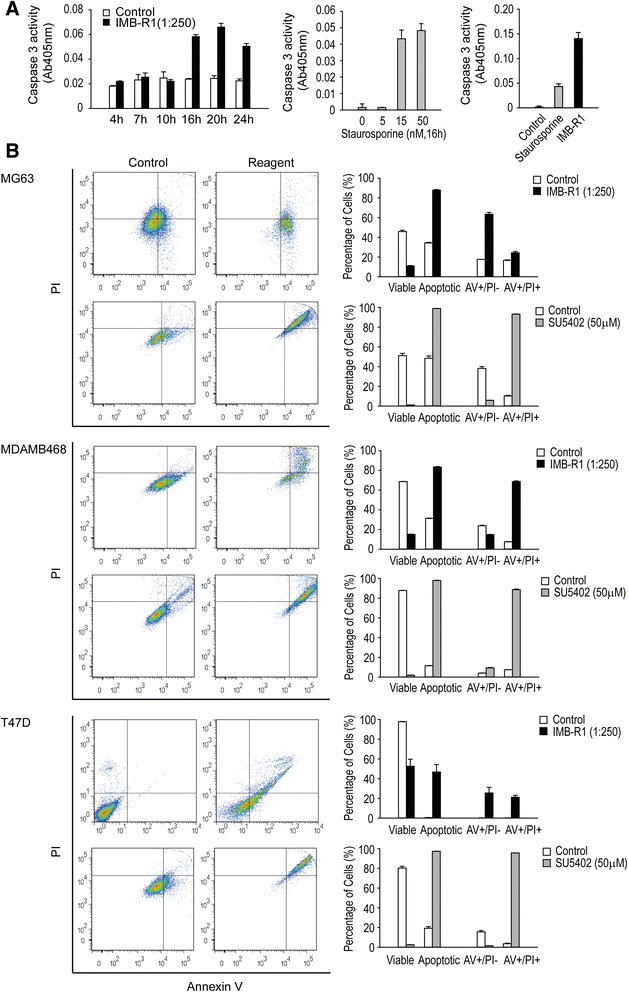
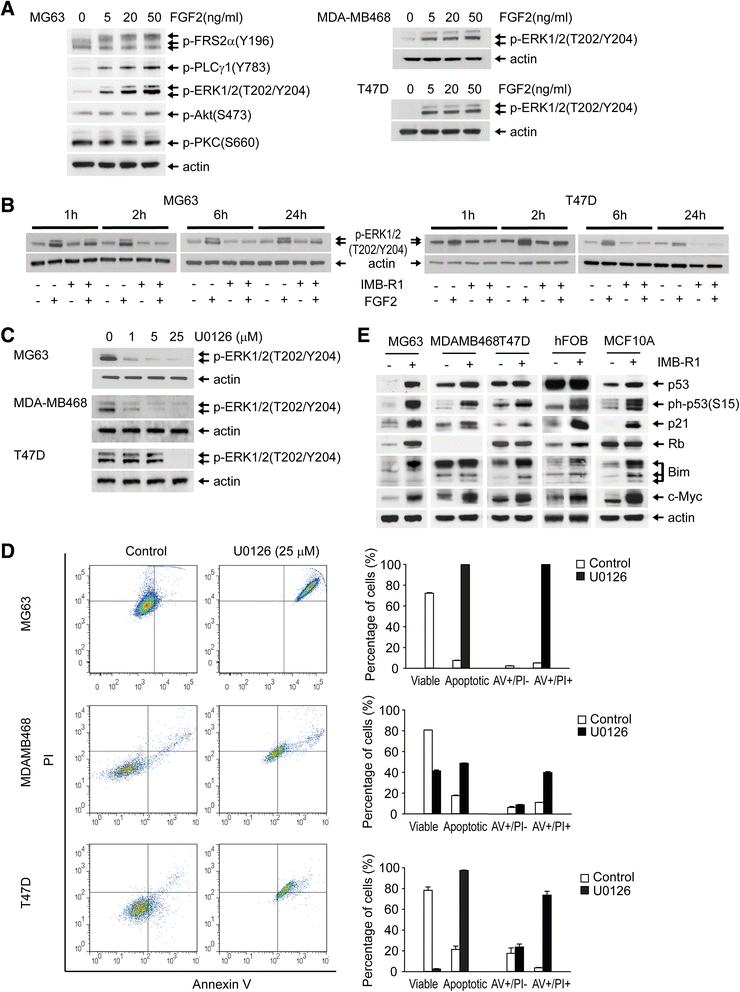
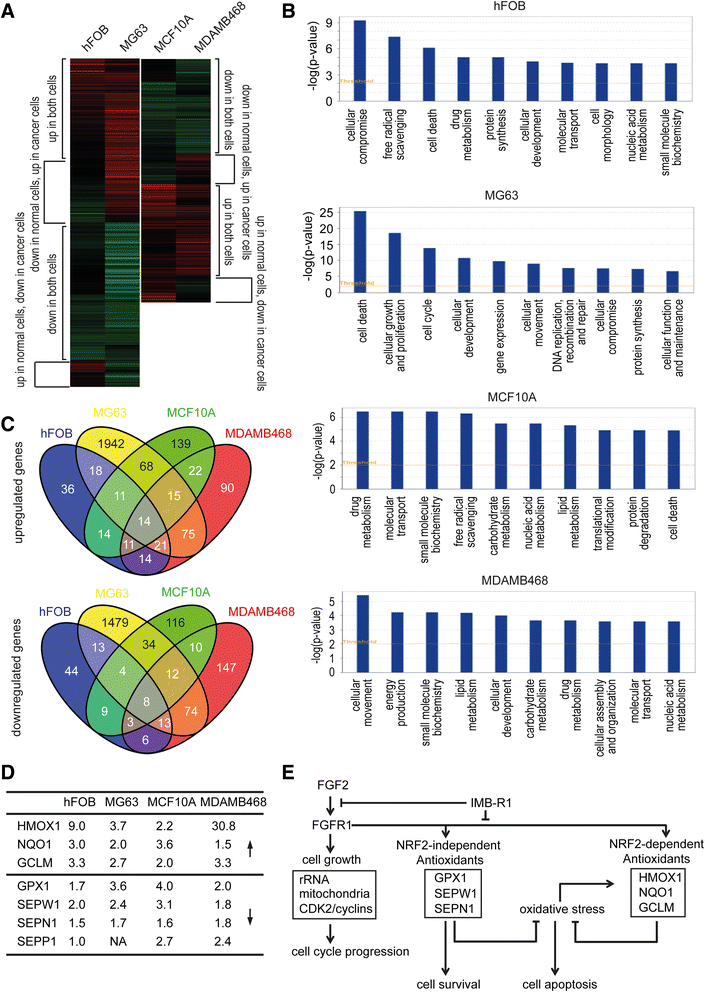
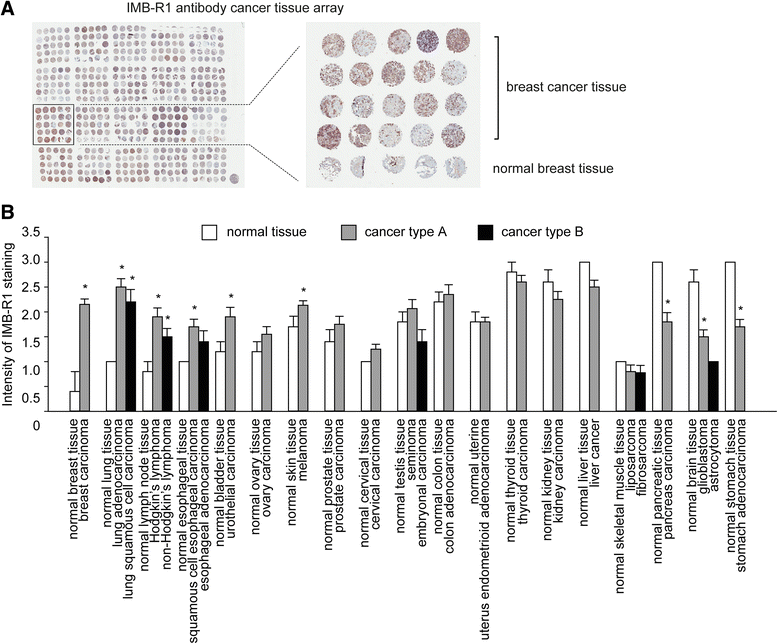
Results: We demonstrate that IMB-R1 is minimally cross-reactive for other FGFRs, and that it potently and specifically inhibits binding of heparin to FGFR1. Furthermore, IMB-R1 blocks the interaction of FGF2 with FGFR1, the kinase activity of FGFR1 and activation of intracellular FGFR signaling. Cancer cells treated with IMB-R1 displayed impaired FGF2 signaling, were unable to grow and instead underwent apoptosis. IMB-R1-induced cell death correlated with a disruption of antioxidative defense networks and increased expression of several tumor suppressors and apoptotic proteins, including p53. Immunostaining with IMB-R1 was stronger in human cancer tissues in which the FGFR1 gene is amplified.
Conclusion: Our study suggests that blocking HS interaction with the heparin-binding domains of FGFR1 inhibited cancer cell growth, which can be an attractive strategy to inactivate cancer-related heparin-binding proteins.
Figures
Similar articles
-
Acidic fibroblast growth factor (FGF-1) and FGF receptor 1 signaling in human Y79 retinoblastoma.Arch Ophthalmol. 2005 Mar;123(3):368-76. doi: 10.1001/archopht.123.3.368. Arch Ophthalmol. 2005. PMID: 15767480
-
Novel FGFR inhibitor ponatinib suppresses the growth of non-small cell lung cancer cells overexpressing FGFR1.Oncol Rep. 2013 Jun;29(6):2181-90. doi: 10.3892/or.2013.2386. Epub 2013 Apr 4. Oncol Rep. 2013. PMID: 23563700
-
The therapeutic potential of a novel non-ATP-competitive fibroblast growth factor receptor 1 inhibitor on gastric cancer.Anticancer Drugs. 2015 Apr;26(4):379-87. doi: 10.1097/CAD.0000000000000195. Anticancer Drugs. 2015. PMID: 25521558
-
FGFR inhibitors: Effects on cancer cells, tumor microenvironment and whole-body homeostasis (Review).Int J Mol Med. 2016 Jul;38(1):3-15. doi: 10.3892/ijmm.2016.2620. Epub 2016 May 31. Int J Mol Med. 2016. PMID: 27245147 Free PMC article. Review.
-
Genomic aberrations in the FGFR pathway: opportunities for targeted therapies in solid tumors.Ann Oncol. 2014 Mar;25(3):552-563. doi: 10.1093/annonc/mdt419. Epub 2013 Nov 20. Ann Oncol. 2014. PMID: 24265351 Free PMC article. Review.
Cited by
-
Pleiotropic Effects of Heparins: From Clinical Applications to Molecular Mechanisms in Hepatocellular Carcinoma.Can J Gastroenterol Hepatol. 2018 Oct 22;2018:7568742. doi: 10.1155/2018/7568742. eCollection 2018. Can J Gastroenterol Hepatol. 2018. PMID: 30425976 Free PMC article. Review.
-
MicroRNA-761 targets FGFR1 to suppress the malignancy of osteosarcoma by deactivating PI3K/Akt pathway.Onco Targets Ther. 2019 Oct 15;12:8501-8513. doi: 10.2147/OTT.S208263. eCollection 2019. Onco Targets Ther. 2019. Retraction in: Onco Targets Ther. 2021 Sep 28;14:4937-4938. doi: 10.2147/OTT.S340637. PMID: 31686864 Free PMC article. Retracted.
-
Exploring the key genes and pathways in enchondromas using a gene expression microarray.Oncotarget. 2017 Jul 4;8(27):43967-43977. doi: 10.18632/oncotarget.16700. Oncotarget. 2017. PMID: 28410203 Free PMC article.
-
Liver-gut axis signaling regulates circadian energy metabolism in shift workers.FASEB J. 2024 Nov 30;38(22):e70203. doi: 10.1096/fj.202402102R. FASEB J. 2024. PMID: 39588921 Free PMC article.
-
Enhancing the Efficacy of Stem Cell Therapy with Glycosaminoglycans.Stem Cell Reports. 2020 Jan 14;14(1):105-121. doi: 10.1016/j.stemcr.2019.12.003. Epub 2020 Jan 2. Stem Cell Reports. 2020. PMID: 31902704 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

