The prognostic value of SUMO1/Sentrin specific peptidase 1 (SENP1) in prostate cancer is limited to ERG-fusion positive tumors lacking PTEN deletion
- PMID: 26202067
- PMCID: PMC4512145
- DOI: 10.1186/s12885-015-1555-8
The prognostic value of SUMO1/Sentrin specific peptidase 1 (SENP1) in prostate cancer is limited to ERG-fusion positive tumors lacking PTEN deletion
Abstract
Background: Posttranscriptional protein modification by SUMOylation plays an important role in tumor development and progression. In the current study we analyzed prevalence and prognostic impact of the de-SUMOylation enzyme SENP1 in prostate cancer.
Methods: SENP1 expression was analyzed by immunohistochemistry on a tissue microarray containing more than 12,400 prostate cancer specimens. Results were compared to tumor phenotype, ERG status, genomic deletions of 3p, 5q, 6q and PTEN, and biochemical recurrence.
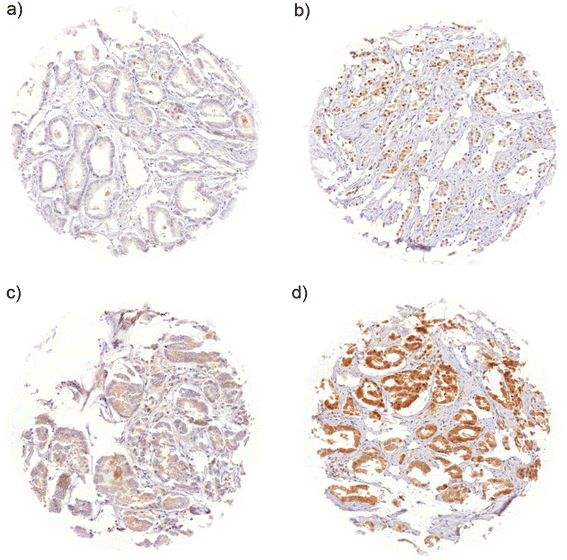
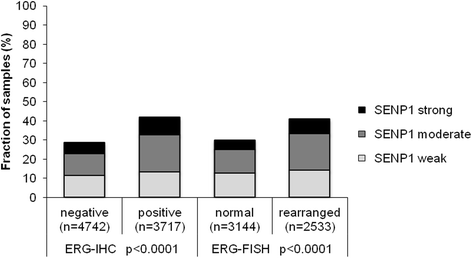
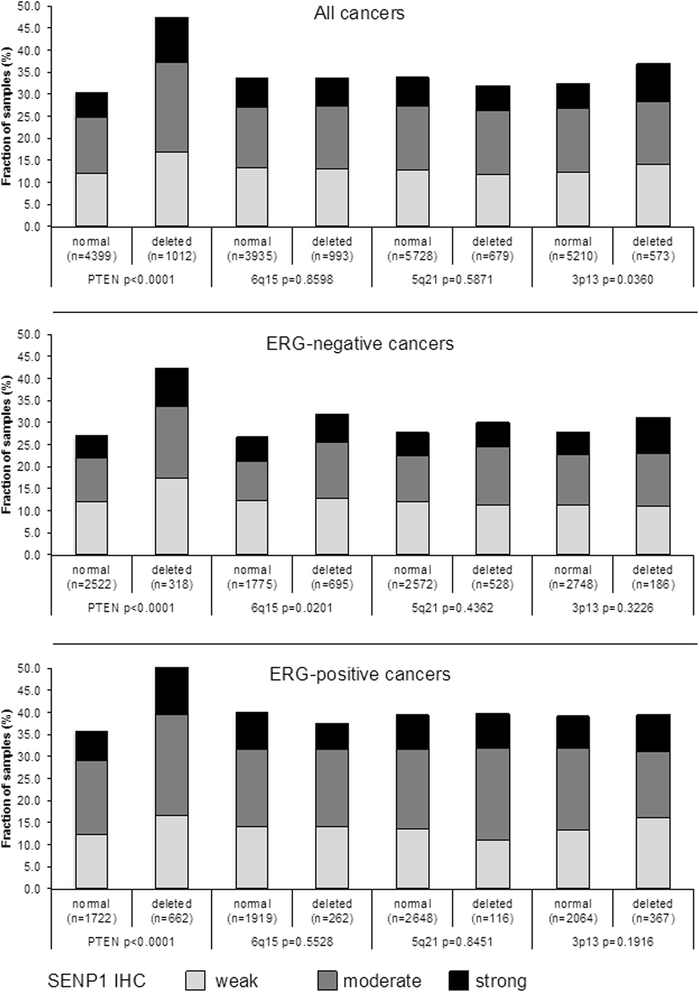
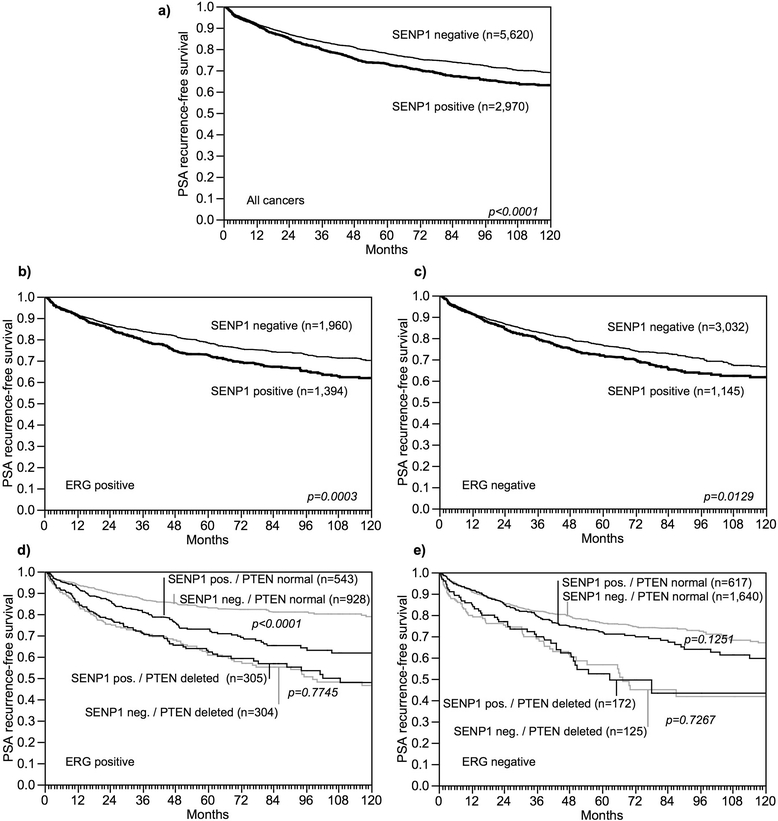
Results: SENP1 immunostaining was detectable in 34.5 % of 9,516 interpretable cancers and considered strong in 7.3 %, moderate in 14.9 % and weak in 12.3 % of cases. Strong SENP1 expression was linked to advanced pT stage (p < 0.0001), high Gleason grade (p < 0.0001), positive lymph node status (p = 0.0019), high pre-operative PSA levels (p = 0.0037), and PSA recurrence (p < 0.0001). SENP1 expression was strongly associated with positive ERG fusion status as determined by both in situ hybridization (FISH) and immunohistochemistry as well as with PTEN deletions. Detectable SENP1 immunostaining was found in 41 % of ERG positive and in 47 % of PTEN deleted cancers but in only 30 % of ERG negative and 30 % of PTEN non-deleted cancers (p < 0.0001 each). Deletions of 3p, 5q, and 6q were unrelated to SENP1 expression. Subset analyses revealed that the prognostic impact of SENP1 expression was solely driven by the subgroup of ERG positive, PTEN undeleted cancers. In this subgroup, the prognostic role of SENP1 expression was independent of the preoperative PSA level, tumor stage, Gleason grade, and the status of the resection margin.
Conclusions: SENP1 expression has strong prognostic impact in a molecularly defined subset of cancers. This is per se not surprising as the biologic impact of each individual molecular event is likely to be dependent on its cellular environment. However, such findings challenge the concept of finding clinically relevant molecular signatures that are equally applicable to all prostate cancers.
Figures
Similar articles
-
βIII-tubulin overexpression is an independent predictor of prostate cancer progression tightly linked to ERG fusion status and PTEN deletion.Am J Pathol. 2014 Mar;184(3):609-17. doi: 10.1016/j.ajpath.2013.11.007. Epub 2013 Dec 28. Am J Pathol. 2014. PMID: 24378408
-
The prognostic impact of high Nijmegen breakage syndrome (NBS1) gene expression in ERG-negative prostate cancers lacking PTEN deletion is driven by KPNA2 expression.Int J Cancer. 2014 Sep 15;135(6):1399-407. doi: 10.1002/ijc.28778. Epub 2014 Feb 26. Int J Cancer. 2014. PMID: 24510842
-
Overexpression of the A Disintegrin and Metalloproteinase ADAM15 is linked to a Small but Highly Aggressive Subset of Prostate Cancers.Neoplasia. 2017 Apr;19(4):279-287. doi: 10.1016/j.neo.2017.01.005. Epub 2017 Mar 8. Neoplasia. 2017. PMID: 28282546 Free PMC article.
-
ERG protein expression as a biomarker of prostate cancer.Biomark Med. 2013 Dec;7(6):851-65. doi: 10.2217/bmm.13.105. Biomark Med. 2013. PMID: 24266818 Review.
-
The prognostic role of ERG immunopositivity in prostatic acinar adenocarcinoma: a study including 454 cases and review of the literature.Hum Pathol. 2014 Mar;45(3):488-97. doi: 10.1016/j.humpath.2013.10.012. Epub 2013 Oct 23. Hum Pathol. 2014. PMID: 24406017 Review.
Cited by
-
High altitude exposure affects male reproductive parameters: could it also affect the prostate?†.Biol Reprod. 2022 Mar 19;106(3):385-396. doi: 10.1093/biolre/ioab205. Biol Reprod. 2022. PMID: 34725677 Free PMC article. Review.
-
Association of PTEN expression with biochemical recurrence in prostate cancer: results based on previous reports.Onco Targets Ther. 2017 Oct 24;10:5089-5097. doi: 10.2147/OTT.S132653. eCollection 2017. Onco Targets Ther. 2017. PMID: 29123407 Free PMC article.
-
A cellular and bioinformatics analysis of the SENP1 SUMO isopeptidase in pancreatic cancer.J Gastrointest Oncol. 2019 Oct;10(5):821-830. doi: 10.21037/jgo.2019.05.09. J Gastrointest Oncol. 2019. PMID: 31602319 Free PMC article.
-
Upregulation of centromere protein F is linked to aggressive prostate cancers.Cancer Manag Res. 2018 Nov 9;10:5491-5504. doi: 10.2147/CMAR.S165630. eCollection 2018. Cancer Manag Res. 2018. PMID: 30519097 Free PMC article.
-
Loss of cytoplasmic survivin expression is an independent predictor of poor prognosis in radically operated prostate cancer patients.Cancer Med. 2020 Feb;9(4):1409-1418. doi: 10.1002/cam4.2773. Epub 2020 Jan 1. Cancer Med. 2020. PMID: 31893572 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

