The Cellular Factor NXP2/MORC3 Is a Positive Regulator of Influenza Virus Multiplication
- PMID: 26202233
- PMCID: PMC4577880
- DOI: 10.1128/JVI.01530-15
The Cellular Factor NXP2/MORC3 Is a Positive Regulator of Influenza Virus Multiplication
Erratum in
-
Correction for Ver et al., "The Cellular Factor NXP2/MORC3 Is a Positive Regulator of Influenza Virus Multiplication".J Virol. 2017 Dec 14;92(1):e01757-17. doi: 10.1128/JVI.01757-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29242254 Free PMC article. No abstract available.
Abstract
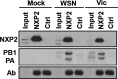
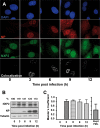
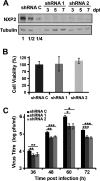
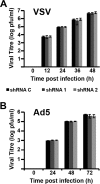
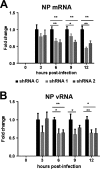
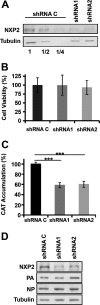
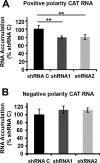
Transcription and replication of influenza A virus are carried out in the nuclei of infected cells in the context of viral ribonucleoproteins (RNPs). The viral polymerase responsible for these processes is a protein complex composed of the PB1, PB2, and PA proteins. We previously identified a set of polymerase-associated cellular proteins by proteomic analysis of polymerase-containing intracellular complexes expressed and purified from human cells. Here we characterize the role of NXP2/MORC3 in the infection cycle. NXP2/MORC3 is a member of the Microrchidia (MORC) family that is associated with the nuclear matrix and has RNA-binding activity. Influenza virus infection led to a slight increase in NXP2/MORC3 expression and its partial relocalization to the cytoplasm. Coimmunoprecipitation and immunofluorescence experiments indicated an association of NXP2/MORC3 with the viral polymerase and RNPs during infection. Downregulation of NXP2/MORC3 by use of two independent short hairpin RNAs (shRNAs) reduced virus titers in low-multiplicity infections. Consistent with these findings, analysis of virus-specific RNA in high-multiplicity infections indicated a reduction of viral RNA (vRNA) and mRNA after NXP2/MORC3 downregulation. Silencing of NXP2/MORC3 in a recombinant minireplicon system in which virus transcription and replication are uncoupled showed reductions in cat mRNA and chloramphenicol acetyltransferase (CAT) protein accumulation but no alterations in cat vRNA levels, suggesting that NXP2/MORC3 is important for influenza virus transcription.
Importance: Influenza virus infections appear as yearly epidemics and occasional pandemics of respiratory disease, with high morbidity and occasional mortality. Influenza viruses are intracellular parasites that replicate and transcribe their genomic ribonucleoproteins in the nuclei of infected cells, in a complex interplay with host cell factors. Here we characterized the role of the human NXP2/MORC3 protein, a member of the Microrchidia family that is associated with the nuclear matrix, during virus infection. NXP2/MORC3 associates with the viral ribonucleoproteins in infected cells. Downregulation of NXP2/MORC3 reduced virus titers and accumulations of viral genomic RNA and mRNAs. Silencing of NXP2/MORC3 in an influenza virus CAT minireplicon system diminished CAT protein and cat mRNA levels but not genomic RNA levels. We propose that NXP2/MORC3 plays a role in influenza virus transcription.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
References
-
- Palese P, Shaw M. 2013. Orthomyxoviridae, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

