Transcriptional Profiling of the Immune Response to Marburg Virus Infection
- PMID: 26202234
- PMCID: PMC4577901
- DOI: 10.1128/JVI.01142-15
Transcriptional Profiling of the Immune Response to Marburg Virus Infection
Abstract
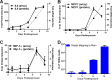
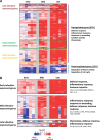
Marburg virus is a genetically simple RNA virus that causes a severe hemorrhagic fever in humans and nonhuman primates. The mechanism of pathogenesis of the infection is not well understood, but it is well accepted that pathogenesis is appreciably driven by a hyperactive immune response. To better understand the overall response to Marburg virus challenge, we undertook a transcriptomic analysis of immune cells circulating in the blood following aerosol exposure of rhesus macaques to a lethal dose of Marburg virus. Using two-color microarrays, we analyzed the transcriptomes of peripheral blood mononuclear cells that were collected throughout the course of infection from 1 to 9 days postexposure, representing the full course of the infection. The response followed a 3-stage induction (early infection, 1 to 3 days postexposure; midinfection, 5 days postexposure; late infection, 7 to 9 days postexposure) that was led by a robust innate immune response. The host response to aerosolized Marburg virus was evident at 1 day postexposure. Analysis of cytokine transcripts that were overexpressed during infection indicated that previously unanalyzed cytokines are likely induced in response to exposure to Marburg virus and further suggested that the early immune response is skewed toward a Th2 response that would hamper the development of an effective antiviral immune response early in disease. Late infection events included the upregulation of coagulation-associated factors. These findings demonstrate very early host responses to Marburg virus infection and provide a rich data set for identification of factors expressed throughout the course of infection that can be investigated as markers of infection and targets for therapy.
Importance: Marburg virus causes a severe infection that is associated with high mortality and hemorrhage. The disease is associated with an immune response that contributes to the lethality of the disease. In this study, we investigated how the immune cells circulating in the blood of infected primates respond following exposure to Marburg virus. Our results show that there are three discernible stages of response to infection that correlate with presymptomatic, early, and late symptomatic stages of infection, a response format similar to that seen following challenge with other hemorrhagic fever viruses. In contrast to the ability of the virus to block innate immune signaling in vitro, the earliest and most sustained response is an interferon-like response. Our analysis also identifies a number of cytokines that are transcriptionally upregulated during late stages of infection and suggest that there is a Th2-skewed response to infection. When correlated with companion data describing the animal model from which our samples were collected, our results suggest that the innate immune response may contribute to overall pathogenesis.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Bausch DG, Nichol ST, Muyembe-Tamfum JJ, Borchert M, Rollin PE, Sleurs H, Campbell P, Tshioko FK, Roth C, Colebunders R, Pirard P, Mardel S, Olinda LA, Zeller H, Tshomba A, Kulidri A, Libande ML, Mulangu S, Formenty P, Grein T, Leirs H, Braack L, Ksiazek T, Zaki S, Bowen MD, Smit SB, Leman PA, Burt FJ, Kemp A, Swanepoel R. 2006. Marburg hemorrhagic fever associated with multiple genetic lineages of virus. N Engl J Med 355:909–919. doi:10.1056/NEJMoa051465. - DOI - PubMed
-
- Towner JS, Khristova ML, Sealy TK, Vincent MJ, Erickson BR, Bawiec DA, Hartman AL, Comer JA, Zaki SR, Stroher U, Gomes da Silva F, del Castillo F, Rollin PE, Ksiazek TG, Nichol ST. 2006. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J Virol 80:6497–6516. doi:10.1128/JVI.00069-06. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

