Molecular Epidemiology and Evolution of Influenza Viruses Circulating within European Swine between 2009 and 2013
- PMID: 26202246
- PMCID: PMC4577897
- DOI: 10.1128/JVI.00840-15
Molecular Epidemiology and Evolution of Influenza Viruses Circulating within European Swine between 2009 and 2013
Abstract
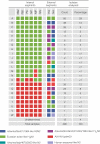
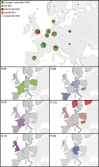
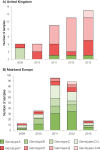
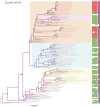
The emergence in humans of the A(H1N1)pdm09 influenza virus, a complex reassortant virus of swine origin, highlighted the importance of worldwide influenza virus surveillance in swine. To date, large-scale surveillance studies have been reported for southern China and North America, but such data have not yet been described for Europe. We report the first large-scale genomic characterization of 290 swine influenza viruses collected from 14 European countries between 2009 and 2013. A total of 23 distinct genotypes were identified, with the 7 most common comprising 82% of the incidence. Contrasting epidemiological dynamics were observed for two of these genotypes, H1huN2 and H3N2, with the former showing multiple long-lived geographically isolated lineages, while the latter had short-lived geographically diffuse lineages. At least 32 human-swine transmission events have resulted in A(H1N1)pdm09 becoming established at a mean frequency of 8% across European countries. Notably, swine in the United Kingdom have largely had a replacement of the endemic Eurasian avian virus-like ("avian-like") genotypes with A(H1N1)pdm09-derived genotypes. The high number of reassortant genotypes observed in European swine, combined with the identification of a genotype similar to the A(H3N2)v genotype in North America, underlines the importance of continued swine surveillance in Europe for the purposes of maintaining public health. This report further reveals that the emergences and drivers of virus evolution in swine differ at the global level.
Importance: The influenza A(H1N1)pdm09 virus contains a reassortant genome with segments derived from separate virus lineages that evolved in different regions of the world. In particular, its neuraminidase and matrix segments were derived from the Eurasian avian virus-like ("avian-like") lineage that emerged in European swine in the 1970s. However, while large-scale genomic characterization of swine has been reported for southern China and North America, no equivalent study has yet been reported for Europe. Surveillance of swine herds across Europe between 2009 and 2013 revealed that the A(H1N1)pdm09 virus is established in European swine, increasing the number of circulating lineages in the region and increasing the possibility of the emergence of a genotype with human pandemic potential. It also has implications for veterinary health, making prevention through vaccination more challenging. The identification of a genotype similar to the A(H3N2)v genotype, causing zoonoses at North American agricultural fairs, underlines the importance of continued genomic characterization in European swine.
Copyright © 2015 Watson et al.
Figures
Similar articles
-
[Swine influenza virus: evolution mechanism and epidemic characterization--a review].Wei Sheng Wu Xue Bao. 2009 Sep;49(9):1138-45. Wei Sheng Wu Xue Bao. 2009. PMID: 20030049 Review. Chinese.
-
Influenza A Viruses of Swine (IAV-S) in Vietnam from 2010 to 2015: Multiple Introductions of A(H1N1)pdm09 Viruses into the Pig Population and Diversifying Genetic Constellations of Enzootic IAV-S.J Virol. 2016 Dec 16;91(1):e01490-16. doi: 10.1128/JVI.01490-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795418 Free PMC article.
-
Genomic diversity and evolutionary dynamics of Influenza A viruses in Colombian swine: implications for one health surveillance and control.Emerg Microbes Infect. 2024 Dec;13(1):2368202. doi: 10.1080/22221751.2024.2368202. Epub 2024 Jul 6. Emerg Microbes Infect. 2024. PMID: 38970562 Free PMC article.
-
Emergence and Evolution of Novel Reassortant Influenza A Viruses in Canines in Southern China.mBio. 2018 Jun 5;9(3):e00909-18. doi: 10.1128/mBio.00909-18. mBio. 2018. PMID: 29871917 Free PMC article.
-
Genetics, evolution, and the zoonotic capacity of European Swine influenza viruses.Curr Top Microbiol Immunol. 2013;370:29-55. doi: 10.1007/82_2012_267. Curr Top Microbiol Immunol. 2013. PMID: 23011571 Review.
Cited by
-
Rapid detection and subtyping of European swine influenza viruses in porcine clinical samples by haemagglutinin- and neuraminidase-specific tetra- and triplex real-time RT-PCRs.Influenza Other Respir Viruses. 2016 Nov;10(6):504-517. doi: 10.1111/irv.12407. Epub 2016 Aug 9. Influenza Other Respir Viruses. 2016. PMID: 27397600 Free PMC article.
-
Human-Origin Influenza A(H3N2) Reassortant Viruses in Swine, Southeast Mexico.Emerg Infect Dis. 2019 Apr;25(4):691-700. doi: 10.3201/eid2504.180779. Epub 2019 Apr 17. Emerg Infect Dis. 2019. PMID: 30730827 Free PMC article.
-
Molecular subtyping of European swine influenza viruses and scaling to high-throughput analysis.Virol J. 2018 Jan 10;15(1):7. doi: 10.1186/s12985-018-0920-z. Virol J. 2018. PMID: 29316958 Free PMC article.
-
The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016.J Gen Virol. 2017 Aug;98(8):2001-2010. doi: 10.1099/jgv.0.000885. Epub 2017 Jul 31. J Gen Virol. 2017. PMID: 28758634 Free PMC article.
-
Influenza A Viruses in the Swine Population: Ecology and Geographical Distribution.Viruses. 2024 Nov 1;16(11):1728. doi: 10.3390/v16111728. Viruses. 2024. PMID: 39599843 Free PMC article. Review.
References
-
- Zell R, Scholtissek C, Ludwig S. 2012. Genetics, evolution, and the zoonotic capacity of European swine influenza viruses, p 29–55. In Richt JA, Webby RJ (ed), Swine influenza. Springer, Berlin, Germany. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

