tRNA recognition by a bacterial tRNA Xm32 modification enzyme from the SPOUT methyltransferase superfamily
- PMID: 26202969
- PMCID: PMC4551947
- DOI: 10.1093/nar/gkv745
tRNA recognition by a bacterial tRNA Xm32 modification enzyme from the SPOUT methyltransferase superfamily
Abstract
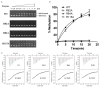
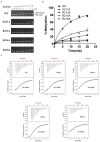
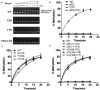
TrmJ proteins from the SPOUT methyltransferase superfamily are tRNA Xm32 modification enzymes that occur in bacteria and archaea. Unlike archaeal TrmJ, bacterial TrmJ require full-length tRNA molecules as substrates. It remains unknown how bacterial TrmJs recognize substrate tRNAs and specifically catalyze a 2'-O modification at ribose 32. Herein, we demonstrate that all six Escherichia coli (Ec) tRNAs with 2'-O-methylated nucleosides at position 32 are substrates of EcTrmJ, and we show that the elbow region of tRNA, but not the amino acid acceptor stem, is needed for the methylation reaction. Our crystallographic study reveals that full-length EcTrmJ forms an unusual dimer in the asymmetric unit, with both the catalytic SPOUT domain and C-terminal extension forming separate dimeric associations. Based on these findings, we used electrophoretic mobility shift assay, isothermal titration calorimetry and enzymatic methods to identify amino acids within EcTrmJ that are involved in tRNA binding. We found that tRNA recognition by EcTrmJ involves the cooperative influences of conserved residues from both the SPOUT and extensional domains, and that this process is regulated by the flexible hinge region that connects these two domains.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





Similar articles
-
Tied up in knots: Untangling substrate recognition by the SPOUT methyltransferases.J Biol Chem. 2022 Oct;298(10):102393. doi: 10.1016/j.jbc.2022.102393. Epub 2022 Aug 18. J Biol Chem. 2022. PMID: 35988649 Free PMC article. Review.
-
Characterization of two homologous 2'-O-methyltransferases showing different specificities for their tRNA substrates.RNA. 2014 Aug;20(8):1257-71. doi: 10.1261/rna.044503.114. Epub 2014 Jun 20. RNA. 2014. PMID: 24951554 Free PMC article.
-
Mechanistic features of the atypical tRNA m1G9 SPOUT methyltransferase, Trm10.Nucleic Acids Res. 2017 Sep 6;45(15):9019-9029. doi: 10.1093/nar/gkx620. Nucleic Acids Res. 2017. PMID: 28911116 Free PMC article.
-
The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase.BMC Mol Biol. 2006 Jul 18;7:23. doi: 10.1186/1471-2199-7-23. BMC Mol Biol. 2006. PMID: 16848900 Free PMC article.
-
Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA.Biomolecules. 2017 Feb 28;7(1):23. doi: 10.3390/biom7010023. Biomolecules. 2017. PMID: 28264529 Free PMC article. Review.
Cited by
-
X-ray structure of the direct electron transfer-type FAD glucose dehydrogenase catalytic subunit complexed with a hitchhiker protein.Acta Crystallogr D Struct Biol. 2019 Sep 1;75(Pt 9):841-851. doi: 10.1107/S2059798319010878. Epub 2019 Aug 28. Acta Crystallogr D Struct Biol. 2019. PMID: 31478907 Free PMC article.
-
Sequence-specific and Shape-selective RNA Recognition by the Human RNA 5-Methylcytosine Methyltransferase NSun6.J Biol Chem. 2016 Nov 11;291(46):24293-24303. doi: 10.1074/jbc.M116.742569. Epub 2016 Oct 4. J Biol Chem. 2016. PMID: 27703015 Free PMC article.
-
Transfer RNA Modification Enzymes with a Thiouridine Synthetase, Methyltransferase and Pseudouridine Synthase (THUMP) Domain and the Nucleosides They Produce in tRNA.Genes (Basel). 2023 Jan 31;14(2):382. doi: 10.3390/genes14020382. Genes (Basel). 2023. PMID: 36833309 Free PMC article. Review.
-
tRNA m1G9 modification depends on substrate-specific RNA conformational changes induced by the methyltransferase Trm10.bioRxiv [Preprint]. 2023 Oct 19:2023.02.01.526536. doi: 10.1101/2023.02.01.526536. bioRxiv. 2023. Update in: J Biol Chem. 2023 Dec;299(12):105443. doi: 10.1016/j.jbc.2023.105443. PMID: 36778341 Free PMC article. Updated. Preprint.
-
Tied up in knots: Untangling substrate recognition by the SPOUT methyltransferases.J Biol Chem. 2022 Oct;298(10):102393. doi: 10.1016/j.jbc.2022.102393. Epub 2022 Aug 18. J Biol Chem. 2022. PMID: 35988649 Free PMC article. Review.
References
-
- El Yacoubi B., Bailly M., de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012;46:69–95. - PubMed
-
- Grosjean H. DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Austin, TX: Landes Bioscience; 2009.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

