Genomic variations of the mevalonate pathway in porokeratosis
- PMID: 26202976
- PMCID: PMC4511816
- DOI: 10.7554/eLife.06322
Genomic variations of the mevalonate pathway in porokeratosis
Erratum in
-
Correction: Genomic variations of the mevalonate pathway in porokeratosis.Elife. 2016 Jan 27;5:e14383. doi: 10.7554/eLife.14383. Elife. 2016. PMID: 26816331 Free PMC article. No abstract available.
Abstract
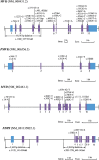
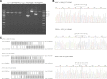
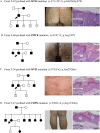
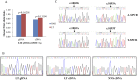
Porokeratosis (PK) is a heterogeneous group of keratinization disorders. No causal genes except MVK have been identified, even though the disease was linked to several genomic loci. Here, we performed massively parallel sequencing and exonic CNV screening of 12 isoprenoid genes in 134 index PK patients (61 familial and 73 sporadic) and identified causal mutations in three novel genes (PMVK, MVD, and FDPS) in addition to MVK in the mevalonate pathway. Allelic expression imbalance (AEI) assays were performed in 13 lesional tissues. At least one mutation in one of the four genes in the mevalonate pathway was found in 60 (98%) familial and 53 (73%) sporadic patients, which suggests that isoprenoid biosynthesis via the mevalonate pathway may play a role in the pathogenesis of PK. Significantly reduced expression of the wild allele was common in lesional tissues due to gene conversion or some other unknown mechanism. A G-to-A RNA editing was observed in one lesional tissue without AEI. In addition, we observed correlations between the mutations in the four mevalonate pathway genes and clinical manifestations in the PK patients, which might support a new and simplified classification of PK under the guidance of genetic testing.
Keywords: genetic testing; human; human biology; medicine; mevalonate pathway; porokeratosis.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures








References
-
- Cui H, Li L, Wang W, Shen J, Yue Z, Zheng X, Zuo X, Liang B, Gao M, Fan X, Yin X, Shen C, Yang C, Zhang C, Zhang X, Sheng Y, Gao J, Zhu Z, Lin D, Zhang A, Wang Z, Liu S, Sun L, Yang S, Cui Y, Zhang X. Exome sequencing identifies SLC17A9 pathogenic gene in two Chinese pedigrees with disseminated superficial actinic porokeratosis. Journal of Medical Genetics. 2014;51:699–704. doi: 10.1136/jmedgenet-2014-102486. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

