The protein BpsB is a poly-β-1,6-N-acetyl-D-glucosamine deacetylase required for biofilm formation in Bordetella bronchiseptica
- PMID: 26203190
- PMCID: PMC4566253
- DOI: 10.1074/jbc.M115.672469
The protein BpsB is a poly-β-1,6-N-acetyl-D-glucosamine deacetylase required for biofilm formation in Bordetella bronchiseptica
Abstract
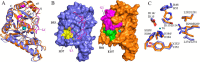
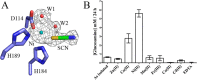
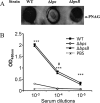
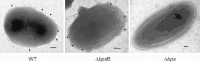
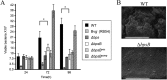
Bordetella pertussis and Bordetella bronchiseptica are the causative agents of whooping cough in humans and a variety of respiratory diseases in animals, respectively. Bordetella species produce an exopolysaccharide, known as the Bordetella polysaccharide (Bps), which is encoded by the bpsABCD operon. Bps is required for Bordetella biofilm formation, colonization of the respiratory tract, and confers protection from complement-mediated killing. In this report, we have investigated the role of BpsB in the biosynthesis of Bps and biofilm formation by B. bronchiseptica. BpsB is a two-domain protein that localizes to the periplasm and outer membrane. BpsB displays metal- and length-dependent deacetylation on poly-β-1,6-N-acetyl-d-glucosamine (PNAG) oligomers, supporting previous immunogenic data that suggests Bps is a PNAG polymer. BpsB can use a variety of divalent metal cations for deacetylase activity and showed highest activity in the presence of Ni(2+) and Co(2+). The structure of the BpsB deacetylase domain is similar to the PNAG deacetylases PgaB and IcaB and contains the same circularly permuted family four carbohydrate esterase motifs. Unlike PgaB from Escherichia coli, BpsB is not required for polymer export and has unique structural differences that allow the N-terminal deacetylase domain to be active when purified in isolation from the C-terminal domain. Our enzymatic characterizations highlight the importance of conserved active site residues in PNAG deacetylation and demonstrate that the C-terminal domain is required for maximal deacetylation of longer PNAG oligomers. Furthermore, we show that BpsB is critical for the formation and complex architecture of B. bronchiseptica biofilms.
Keywords: Bordetella; Bps; Exopolysaccharide Biosynthesis; PNAG; biofilm; carbohydrate esterase; carbohydrate processing; enzyme structure; microbiology; structural biology.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

