The Genera of Fungi - fixing the application of the type species of generic names - G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia
- PMID: 26203422
- PMCID: PMC4500082
- DOI: 10.5598/imafungus.2015.06.01.11
The Genera of Fungi - fixing the application of the type species of generic names - G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia
Abstract
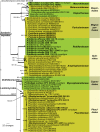
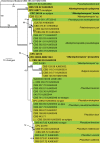
The present paper represents the second contribution in the Genera of Fungi series, linking type species of fungal genera to their morphology and DNA sequence data, and where possible, ecology. This paper focuses on 12 genera of microfungi, 11 of which the type species are neo- or epitypified here: Allantophomopsis (A. cytisporea, Phacidiaceae, Phacidiales, Leotiomycetes), Latorua gen. nov. (Latorua caligans, Latoruaceae, Pleosporales, Dothideomycetes), Macrodiplodiopsis (M. desmazieri, Macrodiplodiopsidaceae, Pleosporales, Dothideomycetes), Macrohilum (M. eucalypti, Macrohilaceae, Diaporthales, Sordariomycetes), Milospium (M. graphideorum, incertae sedis, Pezizomycotina), Protostegia (P. eucleae, Mycosphaerellaceae, Capnodiales, Dothideomycetes), Pyricularia (P. grisea, Pyriculariaceae, Magnaporthales, Sordariomycetes), Robillarda (R. sessilis, Robillardaceae, Xylariales, Sordariomycetes), Rutola (R. graminis, incertae sedis, Pleosporales, Dothideomycetes), Septoriella (S. phragmitis, Phaeosphaeriaceae, Pleosporales, Dothideomycetes), Torula (T. herbarum, Torulaceae, Pleosporales, Dothideomycetes) and Wojnowicia (syn. of Septoriella, S. hirta, Phaeosphaeriaceae, Pleosporales, Dothideomycetes). Novel species include Latorua grootfonteinensis, Robillarda africana, R. roystoneae, R. terrae, Torula ficus, T. hollandica, and T. masonii spp. nov., and three new families: Macrodiplodiopsisceae, Macrohilaceae, and Robillardaceae. Authors interested in contributing accounts of individual genera to larger multi-authored papers to be published in IMA Fungus, should contact the associate editors listed for the major groups of fungi on the List of Protected Generic Names for Fungi (www.generaoffungi.org).
Keywords: DNA Barcodes; ITS; LSU; epitype; fungal systematics; typification; www.GeneraofFungi.org.
Figures












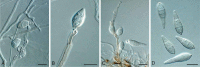






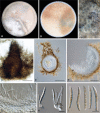






Similar articles
-
The Genera of Fungi - G5: Arthrinium, Ceratosphaeria, Dimerosporiopsis, Hormodochis, Lecanostictopsis, Lembosina, Neomelanconium, Phragmotrichum, Pseudomelanconium, Rutola, and Trullula.Fungal Syst Evol. 2020 Jun;5:77-98. doi: 10.3114/fuse.2020.05.04. Epub 2019 Aug 22. Fungal Syst Evol. 2020. PMID: 32467915 Free PMC article.
-
The Genera of Fungi: fixing the application of type species of generic names.IMA Fungus. 2014 Jun;5(1):141-60. doi: 10.5598/imafungus.2014.05.01.14. Epub 2014 Jun 19. IMA Fungus. 2014. PMID: 25083414 Free PMC article.
-
The Genera of Fungi - G 4: Camarosporium and Dothiora.IMA Fungus. 2017 Jun;8(1):131-152. doi: 10.5598/imafungus.2017.08.01.10. Epub 2017 May 23. IMA Fungus. 2017. PMID: 28824845 Free PMC article.
-
Pyricularia oryzae: Lab star and field scourge.Mol Plant Pathol. 2024 Apr;25(4):e13449. doi: 10.1111/mpp.13449. Mol Plant Pathol. 2024. PMID: 38619508 Free PMC article. Review.
-
Fungal perylenequinones.Mycol Prog. 2022;21(3):38. doi: 10.1007/s11557-022-01790-4. Epub 2022 Apr 4. Mycol Prog. 2022. PMID: 35401071 Free PMC article. Review.
Cited by
-
Foliar pathogens of eucalypts.Stud Mycol. 2019 Aug 8;94:125-298. doi: 10.1016/j.simyco.2019.08.001. eCollection 2019 Sep. Stud Mycol. 2019. PMID: 31636729 Free PMC article.
-
Fungi Present in Antarctic Deep-Sea Sediments Assessed Using DNA Metabarcoding.Microb Ecol. 2021 Jul;82(1):157-164. doi: 10.1007/s00248-020-01658-8. Epub 2021 Jan 6. Microb Ecol. 2021. PMID: 33404819
-
Fungal diversity notes 1512-1610: taxonomic and phylogenetic contributions on genera and species of fungal taxa.Fungal Divers. 2022;117(1):1-272. doi: 10.1007/s13225-022-00513-0. Epub 2023 Feb 23. Fungal Divers. 2022. PMID: 36852303 Free PMC article.
-
Neostagonosporellasichuanensis gen. et sp. nov. (Phaeosphaeriaceae, Pleosporales) on Phyllostachysheteroclada (Poaceae) from Sichuan Province, China.MycoKeys. 2019 Feb 18;(46):119-150. doi: 10.3897/mycokeys.46.32458. eCollection 2019. MycoKeys. 2019. PMID: 30814907 Free PMC article.
-
Families of Diaporthales based on morphological and phylogenetic evidence.Stud Mycol. 2017 Mar;86:217-296. doi: 10.1016/j.simyco.2017.07.003. Epub 2017 Aug 1. Stud Mycol. 2017. PMID: 28947840 Free PMC article.
References
-
- Carris LM. (1990) Cranberry black rot fungi: Allantophomopsis cytisporea and Allantophomopsis lycopodina. Canadian Journal of Botany 68: 2283–2291.
-
- Clements FE, Shear CL. (1931) The Genera of Fungi. New York: H.W. Wilson.
-
- Crane JL, Schoknecht JD. (1977) Revision of Torula species. Rutola, a new genus for Torula graminis. Canadian Journal of Botany 55: 3013–3019.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
