Basidioascus undulatus: genome, origins, and sexuality
- PMID: 26203425
- PMCID: PMC4500085
- DOI: 10.5598/imafungus.2015.06.01.14
Basidioascus undulatus: genome, origins, and sexuality
Abstract
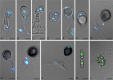
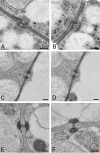
Basidioascus undulatus is a soil basidiomycete belonging to the order Geminibasidiales. The taxonomic status of the order was unclear as originally it was only tentatively classified in the class Wallemiomycetes. The fungi in Geminibasidiales have an ambiguously defined sexual cycle. In this study, we sequenced the genome of B. undulatus to gain insights into its sexuality and evolutionary origins. The assembled genome draft was approximately 32 Mb in size, had a median nucleotide coverage of 24X, and contained 6123 predicted genes. Previous morphological descriptions of B. undulatus relied on interpretation of putative sexual structures. In this study, nuclear staining and confocal microscopy showed meiosis occurring in basidia and genome analysis confirmed the existence of genes involved in meiosis and mating. Using 35 protein-coding genes extracted from genomic information, phylogenomic and molecular dating analyses confirmed that B. undulatus indeed belongs to a lineage distantly related to Wallemia while retaining a basal position in Agaricomycotina. These results, combined with differences in septal pore morphology, led us to move the order Geminibasidiales out of the Wallemiomycetes and into the new class Geminibasidiomycetes cl. nov. Finally, the concept of Agaricomycotina is emended to include both Wallemiomycetes and Geminibasidiomycetes.
Keywords: Agaricomycotina; Basidiomycota; Geminibasidiomycetes; Wallemiomycetes; septal pore ultastructure.
Figures


Similar articles
-
Basidioascus and Geminibasidium: a new lineage of heat-resistant and xerotolerant basidiomycetes.Mycologia. 2013 Sep-Oct;105(5):1231-50. doi: 10.3852/12-351. Epub 2013 May 25. Mycologia. 2013. PMID: 23709525
-
The genome of the xerotolerant mold Wallemia sebi reveals adaptations to osmotic stress and suggests cryptic sexual reproduction.Fungal Genet Biol. 2012 Mar;49(3):217-26. doi: 10.1016/j.fgb.2012.01.007. Epub 2012 Feb 4. Fungal Genet Biol. 2012. PMID: 22326418
-
Basidioascus persicus sp. nov., a yeast-like species of the order Geminibasidiales isolated from soil.Int J Syst Evol Microbiol. 2014 Sep;64(Pt 9):3046-3052. doi: 10.1099/ijs.0.062380-0. Epub 2014 Jun 13. Int J Syst Evol Microbiol. 2014. PMID: 24928426
-
Basidiomycete mating type genes and pheromone signaling.Eukaryot Cell. 2010 Jun;9(6):847-59. doi: 10.1128/EC.00319-09. Epub 2010 Feb 26. Eukaryot Cell. 2010. PMID: 20190072 Free PMC article. Review.
-
A phylogenetic overview of the Agaricomycotina.Mycologia. 2006 Nov-Dec;98(6):917-25. doi: 10.3852/mycologia.98.6.917. Mycologia. 2006. PMID: 17486968 Review.
Cited by
-
Plant part and a steep environmental gradient predict plant microbial composition in a tropical watershed.ISME J. 2021 Apr;15(4):999-1009. doi: 10.1038/s41396-020-00826-5. Epub 2020 Nov 13. ISME J. 2021. PMID: 33188299 Free PMC article.
-
Fungal evolution: diversity, taxonomy and phylogeny of the Fungi.Biol Rev Camb Philos Soc. 2019 Dec;94(6):2101-2137. doi: 10.1111/brv.12550. Biol Rev Camb Philos Soc. 2019. PMID: 31659870 Free PMC article.
-
Microbes affected the TYLCCNV transmission rate by the Q biotype whitefly under high O3.Sci Rep. 2017 Oct 31;7(1):14412. doi: 10.1038/s41598-017-14023-6. Sci Rep. 2017. PMID: 29089507 Free PMC article.
References
-
- Bauer R, Begerow B, Sampaio JP, Weiss M, Oberwinkler F. (2006) The simple-septate basidiomycetes: a synopsis. Mycological Progress 5: 41–66.
-
- Berbee ML, Taylor JW. (2010) Dating the molecular clock in fungi – how close are we? Fungal Biology Reviews 24: 1–16.
-
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. (2011) Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27: 578–579. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
