Genome-editing tools for stem cell biology
- PMID: 26203860
- PMCID: PMC4650720
- DOI: 10.1038/cddis.2015.167
Genome-editing tools for stem cell biology
Abstract
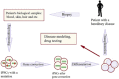
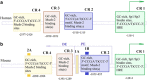
Human pluripotent stem cells provide a versatile platform for regenerative studies, drug testing and disease modeling. That the expression of only four transcription factors, Oct4, Klf4, Sox2 and c-Myc (OKSM), is sufficient for generation of induced pluripotent stem cells (iPSCs) from differentiated somatic cells has revolutionized the field and also highlighted the importance of OKSM as targets for genome editing. A number of novel genome-editing systems have been developed recently. In this review, we focus on successful applications of several such systems for generation of iPSCs. In particular, we discuss genome-editing systems based on zinc-finger fusion proteins (ZFs), transcription activator-like effectors (TALEs) and an RNA-guided DNA-specific nuclease, Cas9, derived from the bacterial defense system against viruses that utilizes clustered regularly interspaced short palindromic repeats (CRISPR).
Figures

References
-
- 1Thomson JA. Embryonic stem cell lines derived from human blastocysts. Science 1998; 282: 1145–1147. - PubMed
-
- 2Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126: 663–676. - PubMed
-
- 3Wei X, Chen Y, Xu Y, Zhan Y, Zhang R, Wang M et al. Small molecule compound induces chromatin de-condensation and facilitates induced pluripotent stem cell generation. J Mol Cell Biol 2014; 6: 409–420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

