The Grapevine VvPMEI1 Gene Encodes a Novel Functional Pectin Methylesterase Inhibitor Associated to Grape Berry Development
- PMID: 26204516
- PMCID: PMC4512722
- DOI: 10.1371/journal.pone.0133810
The Grapevine VvPMEI1 Gene Encodes a Novel Functional Pectin Methylesterase Inhibitor Associated to Grape Berry Development
Abstract
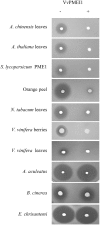
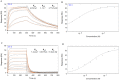
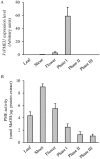
Pectin is secreted in a highly methylesterified form and partially de-methylesterified in the cell wall by pectin methylesterases (PMEs). PME activity is expressed during plant growth, development and stress responses. PME activity is controlled at the post-transcriptional level by proteins named PME inhibitors (PMEIs). We have identified, expressed and characterized VvPMEI1, a functional PME inhibitor of Vitis vinifera. VvPMEI1 typically affects the activity of plant PMEs and is inactive against microbial PMEs. The kinetics of PMEI-PME interaction, studied by surface plasmon resonance, indicates that the inhibitor strongly interacts with PME at apoplastic pH while the stability of the complex is reduced by increasing the pH. The analysis of VvPMEI1 expression in different grapevine tissues and during grape fruit development suggests that this inhibitor controls PME activity mainly during the earlier phase of berry development. A proteomic analysis performed at this stage indicates a PME isoform as possible target of VvPMEI1.
Conflict of interest statement
Figures

Similar articles
-
Genome-Wide Identification and Expression Profiling of the Polygalacturonase (PG) and Pectin Methylesterase (PME) Genes in Grapevine (Vitis vinifera L.).Int J Mol Sci. 2019 Jun 28;20(13):3180. doi: 10.3390/ijms20133180. Int J Mol Sci. 2019. PMID: 31261768 Free PMC article.
-
The Multifaceted Role of Pectin Methylesterase Inhibitors (PMEIs).Int J Mol Sci. 2018 Sep 21;19(10):2878. doi: 10.3390/ijms19102878. Int J Mol Sci. 2018. PMID: 30248977 Free PMC article. Review.
-
Characterization and transcript profiling of the pectin methylesterase (PME) and pectin methylesterase inhibitor (PMEI) gene families in flax (Linum usitatissimum).BMC Genomics. 2013 Oct 30;14:742. doi: 10.1186/1471-2164-14-742. BMC Genomics. 2013. PMID: 24168262 Free PMC article.
-
A functional pectin methylesterase inhibitor protein (SolyPMEI) is expressed during tomato fruit ripening and interacts with PME-1.Plant Mol Biol. 2012 Jul;79(4-5):429-42. doi: 10.1007/s11103-012-9921-2. Epub 2012 May 19. Plant Mol Biol. 2012. PMID: 22610346
-
Pectin methylesterase inhibitor.Biochim Biophys Acta. 2004 Feb 12;1696(2):245-52. doi: 10.1016/j.bbapap.2003.08.011. Biochim Biophys Acta. 2004. PMID: 14871665 Review.
Cited by
-
Genome-Wide Identification, Molecular Evolution, and Expression Profiling Analysis of Pectin Methylesterase Inhibitor Genes in Brassica campestris ssp. chinensis.Int J Mol Sci. 2018 May 2;19(5):1338. doi: 10.3390/ijms19051338. Int J Mol Sci. 2018. PMID: 29724020 Free PMC article.
-
Three Pectin Methylesterase Inhibitors Protect Cell Wall Integrity for Arabidopsis Immunity to Botrytis.Plant Physiol. 2017 Mar;173(3):1844-1863. doi: 10.1104/pp.16.01185. Epub 2017 Jan 12. Plant Physiol. 2017. PMID: 28082716 Free PMC article.
-
Phenolic, Polysaccharides Composition, and Texture Properties during Ripening and Storage Time of New Table Grape Cultivars in Chile.Plants (Basel). 2023 Jun 29;12(13):2488. doi: 10.3390/plants12132488. Plants (Basel). 2023. PMID: 37447049 Free PMC article.
-
Pectin methylesterase gene AtPMEPCRA contributes to physiological adaptation to simulated and spaceflight microgravity in Arabidopsis.iScience. 2022 Apr 29;25(5):104331. doi: 10.1016/j.isci.2022.104331. eCollection 2022 May 20. iScience. 2022. PMID: 35602950 Free PMC article.
-
Genome-Wide Identification, Characterization and Expression Patterns of the Pectin Methylesterase Inhibitor Genes in Sorghum bicolor.Genes (Basel). 2019 Sep 26;10(10):755. doi: 10.3390/genes10100755. Genes (Basel). 2019. PMID: 31561536 Free PMC article.
References
-
- Mohnen D. Pectin structure and biosynthesis. Curr Op Plant Biol.2008; 11: 266–277. - PubMed
-
- Raiola A, Lionetti V, Elmaghraby I, Immerzeel P, Mellerowicz EJ, Salvi G, et al. Pectin methylesterase is induced in Arabidopsis upon infection and is necessary for a successful colonization by necrotrophic pathogens. Mol Plant-Microbe Interact.2011; 24: 432–440. 10.1094/MPMI-07-10-0157 - DOI - PubMed
-
- Ridley BL, O'Neill MA, Mohnen D. Pectins: structure, biosynthesis, and oligogalacturonide-related signaling. Phytochem.2001; 57: 929–967. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

