Abundant copy-number loss of CYCLOPS and STOP genes in gastric adenocarcinoma
- PMID: 26205786
- PMCID: PMC4824836
- DOI: 10.1007/s10120-015-0514-z
Abundant copy-number loss of CYCLOPS and STOP genes in gastric adenocarcinoma
Abstract
Background: Gastric cancer, a leading cause of cancer death worldwide, has been little studied compared with other cancers that impose similar health burdens. Our goal is to assess genomic copy-number loss and the possible functional consequences and therapeutic implications thereof across a large series of gastric adenocarcinomas.
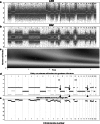
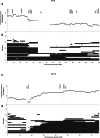
Methods: We used high-density single-nucleotide polymorphism microarrays to determine patterns of copy-number loss and allelic imbalance in 74 gastric adenocarcinomas. We investigated whether suppressor of tumorigenesis and/or proliferation (STOP) genes are associated with genomic copy-number loss. We also analyzed the extent to which copy-number loss affects Copy-number alterations Yielding Cancer Liabilities Owing to Partial losS (CYCLOPS) genes-genes that may be attractive targets for therapeutic inhibition when partially deleted.
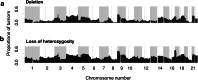
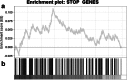
Results: The proportion of the genome subject to copy-number loss varies considerably from tumor to tumor, with a median of 5.5 %, and a mean of 12 % (range 0-58.5 %). On average, 91 STOP genes were subject to copy-number loss per tumor (median 35, range 0-452), and STOP genes tended to have lower copy-number compared with the rest of the genes. Furthermore, on average, 1.6 CYCLOPS genes per tumor were both subject to copy-number loss and downregulated, and 51.4 % of the tumors had at least one such gene.
Conclusions: The enrichment of STOP genes in regions of copy-number loss indicates that their deletion may contribute to gastric carcinogenesis. Furthermore, the presence of several deleted and downregulated CYCLOPS genes in some tumors suggests potential therapeutic targets in these tumors.
Keywords: DNA copy number change; Gastric cancer; Loss of heterozygosity; Tumor suppressor genes.
Figures
References
-
- Uchino S, Tsuda H, Noguchi M, Yokota J, Terada M, Saito T, et al. Frequent loss of heterozygosity at the DCC locus in gastric cancer. Cancer Res. 1992;52(11):3099–3102. - PubMed
-
- Sano T, Tsujino T, Yoshida K, Nakayama H, Haruma K, Ito H, et al. Frequent loss of heterozygosity on chromosomes 1q, 5q, and 17p in human gastric carcinomas. Cancer Res. 1991;51(11):2926–2931. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

