MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures
- PMID: 26205790
- PMCID: PMC4525211
- DOI: 10.1038/ncomms8743
MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures
Erratum in
-
Author Correction: MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures.Nat Commun. 2019 Nov 21;10(1):5290. doi: 10.1038/s41467-019-13200-7. Nat Commun. 2019. PMID: 31754097 Free PMC article.
Abstract
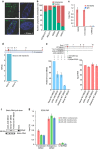
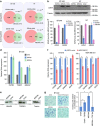
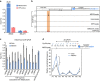
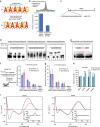
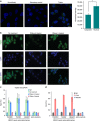
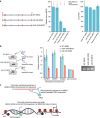
Long noncoding RNAs (lncRNAs) regulate gene expression by association with chromatin, but how they target chromatin remains poorly understood. We have used chromatin RNA immunoprecipitation-coupled high-throughput sequencing to identify 276 lncRNAs enriched in repressive chromatin from breast cancer cells. Using one of the chromatin-interacting lncRNAs, MEG3, we explore the mechanisms by which lncRNAs target chromatin. Here we show that MEG3 and EZH2 share common target genes, including the TGF-β pathway genes. Genome-wide mapping of MEG3 binding sites reveals that MEG3 modulates the activity of TGF-β genes by binding to distal regulatory elements. MEG3 binding sites have GA-rich sequences, which guide MEG3 to the chromatin through RNA-DNA triplex formation. We have found that RNA-DNA triplex structures are widespread and are present over the MEG3 binding sites associated with the TGF-β pathway genes. Our findings suggest that RNA-DNA triplex formation could be a general characteristic of target gene recognition by the chromatin-interacting lncRNAs.
Figures

References
-
- Mohammad F., Mondal T., Guseva N., Pandey G. K. & Kanduri C. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137, 2493–2499 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

