Pseudomonas syringae Effector Avirulence Protein E Localizes to the Host Plasma Membrane and Down-Regulates the Expression of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE13 Gene Required for Antibacterial Immunity in Arabidopsis
- PMID: 26206852
- PMCID: PMC4577396
- DOI: 10.1104/pp.15.00547
Pseudomonas syringae Effector Avirulence Protein E Localizes to the Host Plasma Membrane and Down-Regulates the Expression of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE13 Gene Required for Antibacterial Immunity in Arabidopsis
Abstract
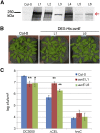
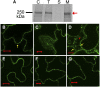
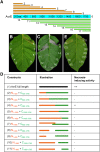
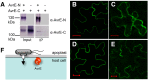
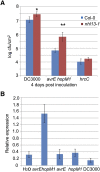
Many bacterial pathogens of plants and animals deliver effector proteins into host cells to promote infection. Elucidation of how pathogen effector proteins function not only is critical for understanding bacterial pathogenesis but also provides a useful tool in discovering the functions of host genes. In this study, we characterized the Pseudomonas syringae pv tomato DC3000 effector protein Avirulence Protein E (AvrE), the founding member of a widely distributed, yet functionally enigmatic, bacterial effector family. We show that AvrE is localized in the plasma membrane (PM) and PM-associated vesicle-like structures in the plant cell. AvrE contains two physically interacting domains, and the amino-terminal portion contains a PM-localization signal. Genome-wide microarray analysis indicates that AvrE, as well as the functionally redundant effector Hypersensitive response and pathogenicity-dependent Outer Protein M1, down-regulates the expression of the NONRACE-SPECIFIC DISEASE RESISTANCE1/HARPIN-INDUCED1-LIKE13 (NHL13) gene in Arabidopsis (Arabidopsis thaliana). Mutational analysis shows that NHL13 is required for plant immunity, as the nhl13 mutant plant displayed enhanced disease susceptibility. Our results defined the action site of one of the most important bacterial virulence proteins in plants and the antibacterial immunity function of the NHL13 gene.
© 2015 American Society of Plant Biologists. All Rights Reserved.
Figures

Similar articles
-
The Erwinia amylovora avrRpt2EA gene contributes to virulence on pear and AvrRpt2EA is recognized by Arabidopsis RPS2 when expressed in pseudomonas syringae.Mol Plant Microbe Interact. 2006 Jun;19(6):644-54. doi: 10.1094/MPMI-19-0644. Mol Plant Microbe Interact. 2006. PMID: 16776298
-
The Pseudomonas syringae type III effector HopD1 suppresses effector-triggered immunity, localizes to the endoplasmic reticulum, and targets the Arabidopsis transcription factor NTL9.New Phytol. 2014 Mar;201(4):1358-1370. doi: 10.1111/nph.12626. Epub 2013 Dec 12. New Phytol. 2014. PMID: 24329768
-
The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana.Plant J. 2007 Nov;52(4):658-72. doi: 10.1111/j.1365-313X.2007.03262.x. Epub 2007 Sep 18. Plant J. 2007. PMID: 17877704
-
Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants.Annu Rev Phytopathol. 2013;51:473-98. doi: 10.1146/annurev-phyto-082712-102321. Epub 2013 May 31. Annu Rev Phytopathol. 2013. PMID: 23725467 Review.
-
Stabbing in the BAK--an original target for avirulence genes of plant pathogens.Cell Host Microbe. 2008 Jul 17;4(1):5-7. doi: 10.1016/j.chom.2008.06.003. Cell Host Microbe. 2008. PMID: 18621005 Review.
Cited by
-
Subcellular Localization of Pseudomonas syringae pv. tomato Effector Proteins in Plants.Methods Mol Biol. 2017;1531:141-153. doi: 10.1007/978-1-4939-6649-3_12. Methods Mol Biol. 2017. PMID: 27837488 Free PMC article.
-
Synergistically promoting plant health by harnessing synthetic microbial communities and prebiotics.iScience. 2021 Jul 30;24(8):102918. doi: 10.1016/j.isci.2021.102918. eCollection 2021 Aug 20. iScience. 2021. PMID: 34430808 Free PMC article. Review.
-
Innovation and Application of the Type III Secretion System Inhibitors in Plant Pathogenic Bacteria.Microorganisms. 2020 Dec 9;8(12):1956. doi: 10.3390/microorganisms8121956. Microorganisms. 2020. PMID: 33317075 Free PMC article. Review.
-
What the Wild Things Do: Mechanisms of Plant Host Manipulation by Bacterial Type III-Secreted Effector Proteins.Microorganisms. 2021 May 11;9(5):1029. doi: 10.3390/microorganisms9051029. Microorganisms. 2021. PMID: 34064647 Free PMC article. Review.
-
Defense against phytopathogens relies on efficient antimicrobial protein secretion mediated by the microtubule-binding protein TGNap1.Nat Commun. 2023 Oct 11;14(1):6357. doi: 10.1038/s41467-023-41807-4. Nat Commun. 2023. PMID: 37821453 Free PMC article.
References
-
- Alfano JR, Charkowski AO, Deng WL, Badel JL, Petnicki-Ocwieja T, van Dijk K, Collmer A (2000) The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci USA 97: 4856–4861 - PMC - PubMed
-
- Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, McMahon SA, Ghosh P, Hughes TR, Boone C, et al. (2006) Identification of a bacterial type III effector family with G protein mimicry functions. Cell 124: 133–145 - PubMed
-
- Badel JL, Shimizu R, Oh HS, Collmer A (2006) A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol Plant Microbe Interact 19: 99–111 - PubMed
-
- Bent A. (2006) Arabidopsis thaliana floral dip transformation method. Methods Mol Biol 343: 87–103 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

