Chlamydia trachomatis In Vivo to In Vitro Transition Reveals Mechanisms of Phase Variation and Down-Regulation of Virulence Factors
- PMID: 26207372
- PMCID: PMC4514472
- DOI: 10.1371/journal.pone.0133420
Chlamydia trachomatis In Vivo to In Vitro Transition Reveals Mechanisms of Phase Variation and Down-Regulation of Virulence Factors
Abstract
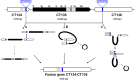
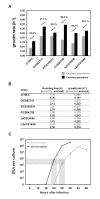
Research on the obligate intracellular bacterium Chlamydia trachomatis demands culture in cell-lines, but the adaptive process behind the in vivo to in vitro transition is not understood. We assessed the genomic and transcriptomic dynamics underlying C. trachomatis in vitro adaptation of strains representing the three disease groups (ocular, epithelial-genital and lymphogranuloma venereum) propagated in epithelial cells over multiple passages. We found genetic features potentially underlying phase variation mechanisms mediating the regulation of a lipid A biosynthesis enzyme (CT533/LpxC), and the functionality of the cytotoxin (CT166) through an ON/OFF mechanism. We detected inactivating mutations in CT713/porB, a scenario suggesting metabolic adaptation to the available carbon source. CT135 was inactivated in a tropism-specific manner, with CT135-negative clones emerging for all epithelial-genital populations (but not for LGV and ocular populations) and rapidly increasing in frequency (~23% mutants per 10 passages). RNA-sequencing analyses revealed that a deletion event involving CT135 impacted the expression of multiple virulence factors, namely effectors known to play a role in the C. trachomatis host-cell invasion or subversion (e.g., CT456/Tarp, CT694, CT875/TepP and CT868/ChlaDub1). This reflects a scenario of attenuation of C. trachomatis virulence in vitro, which may take place independently or in a cumulative fashion with the also observed down-regulation of plasmid-related virulence factors. This issue may be relevant on behalf of the recent advances in Chlamydia mutagenesis and transformation where culture propagation for selecting mutants/transformants is mandatory. Finally, there was an increase in the growth rate for all strains, reflecting gradual fitness enhancement over time. In general, these data shed light on the adaptive process underlying the C. trachomatis in vivo to in vitro transition, and indicates that it would be prudent to restrict culture propagation to minimal passages and check the status of the CT135 genotype in order to avoid the selection of CT135-negative mutants, likely originating less virulent strains.
Conflict of interest statement
Figures




References
-
- Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, Gao Y, et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009; 10:R118 10.1186/gb-2009-10-10-r118 - DOI - PMC - PubMed
-
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003; 4:457–469. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

