A Genomic, Transcriptomic and Proteomic Look at the GE2270 Producer Planobispora rosea, an Uncommon Actinomycete
- PMID: 26207753
- PMCID: PMC4514598
- DOI: 10.1371/journal.pone.0133705
A Genomic, Transcriptomic and Proteomic Look at the GE2270 Producer Planobispora rosea, an Uncommon Actinomycete
Abstract
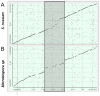
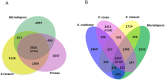
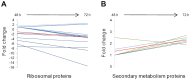
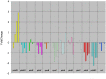
We report the genome sequence of Planobispora rosea ATCC 53733, a mycelium-forming soil-dweller belonging to one of the lesser studied genera of Actinobacteria and producing the thiopeptide GE2270. The P. rosea genome presents considerable convergence in gene organization and function with other members in the family Streptosporangiaceae, with a significant number (44%) of shared orthologs. Patterns of gene expression in P. rosea cultures during exponential and stationary phase have been analyzed using whole transcriptome shotgun sequencing and by proteome analysis. Among the differentially abundant proteins, those involved in protein metabolism are particularly represented, including the GE2270-insensitive EF-Tu. Two proteins from the pbt cluster, directing GE2270 biosynthesis, slightly increase their abundance values over time. While GE2270 production starts during the exponential phase, most pbt genes, as analyzed by qRT-PCR, are down-regulated. The exception is represented by pbtA, encoding the precursor peptide of the ribosomally synthesized GE2270, whose expression reached the highest level at the entry into stationary phase.
Conflict of interest statement
Figures



References
-
- Vobis G, Lodders N., Kämpfer P. Genus VII.Planobispora—The Actinobacteria, Part B In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo ME, Suzuki KI, Ludwig W, et al., editors. Bergey's Manual of Systematic Bacteriology. 5 Second ed. New York: Springer; 2012. pp. 1861–6.
-
- Thiemann JE, Beretta G. A new genus of the Actinoplanaceae: Planobispora, gen. nov. Archiv fur Mikrobiologie. 1968;62: 157–66. Epub 1968/01/01. . - PubMed
-
- Goodfellow M, Stanton LJ, Simpson KE, Minnikin DE. Numerical and chemical classification of Actinoplanes and some related actinomycetes. Journal of General Microbiology. 1990;136: 19–36. 10.1099/00221287-136-1-19 - DOI
-
- Thiemann JE. Studies of Some new genera and species of the Actinoplanaceae In: H. P, editor. The Actinomycetales. Jena, Germany: Gustav Fischer Verlag; 1970. p. 245–57.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

