Agile text mining for the 2014 i2b2/UTHealth Cardiac risk factors challenge
- PMID: 26209007
- PMCID: PMC4737484
- DOI: 10.1016/j.jbi.2015.06.030
Agile text mining for the 2014 i2b2/UTHealth Cardiac risk factors challenge
Abstract
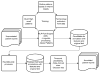
This paper describes the use of an agile text mining platform (Linguamatics' Interactive Information Extraction Platform, I2E) to extract document-level cardiac risk factors in patient records as defined in the i2b2/UTHealth 2014 challenge. The approach uses a data-driven rule-based methodology with the addition of a simple supervised classifier. We demonstrate that agile text mining allows for rapid optimization of extraction strategies, while post-processing can leverage annotation guidelines, corpus statistics and logic inferred from the gold standard data. We also show how data imbalance in a training set affects performance. Evaluation of this approach on the test data gave an F-Score of 91.7%, one percent behind the top performing system.
Keywords: Clinical natural language processing; Information extraction; Text mining.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures





References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

