Molecular studies on ancient M. tuberculosis and M. leprae: methods of pathogen and host DNA analysis
- PMID: 26210385
- PMCID: PMC4545183
- DOI: 10.1007/s10096-015-2427-5
Molecular studies on ancient M. tuberculosis and M. leprae: methods of pathogen and host DNA analysis
Abstract
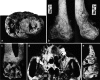
Humans have evolved alongside infectious diseases for millennia. Despite the efforts to reduce their incidence, infectious diseases still pose a tremendous threat to the world population. Fast development of molecular techniques and increasing risk of new epidemics have resulted in several studies that look to the past in order to investigate the origin and evolution of infectious diseases. Tuberculosis and leprosy have become frequent targets of such studies, owing to the persistence of their molecular biomarkers in ancient material and the characteristic skeletal lesions each disease may cause. This review examines the molecular methods used to screen for the presence of M. tuberculosis and M. leprae ancient DNA (aDNA) and their differentiation in ancient human remains. Examples of recent studies, mainly from Europe, that employ the newest techniques of molecular analysis are also described. Moreover, we present a specific approach based on assessing the likely immunological profile of historic populations, in order to further elucidate the influence of M. tuberculosis and M. leprae on historical human populations.
Figures

References
-
- Roberts CA, Buikstra JE. The bioarchaeology of tuberculosis: a global view on a reemerging disease. Gainsville: University Press of Florida; 2003.
-
- Ortner DJ. Identification of pathological conditions in human skeletal remains. Amsterdam: Academic Press; 2003.
-
- Gładykowska-Rzeczycka J. Sources, methods and documentation in paleopathology and archeopathology. In: Dzieduszycki W, Wrzesiński J, editors. Methods, sources, documentation. Funeralia at Lednica. Poznań: Scientific Association of Polish Archaeologists, Poznań Branch; 2009. pp. 83–92.
-
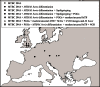
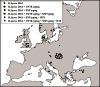
- Donoghue HD, Michael Taylor G, Marcsik A, Molnár E, Pálfi G, Pap I, Teschler-Nicola M, Pinhasi R, Erdal YS, Velemínsky P, Likovsky J, Belcastro MG, Mariotti V, Riga A, Rubini M, Zaio P, Besra GS, Lee OY, Wu HH, Minnikin DE, Bull ID, O’Grady J, Spigelman M. A migration-driven model for the historical spread of leprosy in medieval Eastern and Central Europe. Infect Genet Evol. 2015;31:250–256. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

