Lymphocyte-specific protein tyrosine kinase (Lck) interacts with CR6-interacting factor 1 (CRIF1) in mitochondria to repress oxidative phosphorylation
- PMID: 26210498
- PMCID: PMC4515320
- DOI: 10.1186/s12885-015-1520-6
Lymphocyte-specific protein tyrosine kinase (Lck) interacts with CR6-interacting factor 1 (CRIF1) in mitochondria to repress oxidative phosphorylation
Abstract
Background: Many cancer cells exhibit reduced mitochondrial respiration as part of metabolic reprogramming to support tumor growth. Mitochondrial localization of several protein tyrosine kinases is linked to this characteristic metabolic shift in solid tumors, but remains largely unknown in blood cancer. Lymphocyte-specific protein tyrosine kinase (Lck) is a key T-cell kinase and widely implicated in blood malignancies. The purpose of our study is to determine whether and how Lck contributes to metabolic shift in T-cell leukemia through mitochondrial localization.
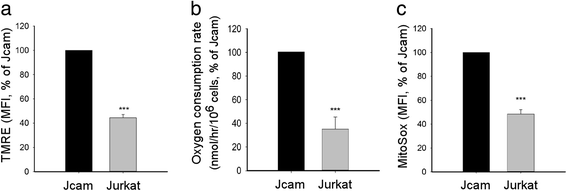
Methods: We compared the human leukemic T-cell line Jurkat with its Lck-deficient derivative Jcam cell line. Differences in mitochondrial respiration were measured by the levels of mitochondrial membrane potential, oxygen consumption, and mitochondrial superoxide. Detailed mitochondrial structure was visualized by transmission electron microscopy. Lck localization was evaluated by subcellular fractionation and confocal microscopy. Proteomic analysis was performed to identify proteins co-precipitated with Lck in leukemic T-cells. Protein interaction was validated by biochemical co-precipitation and confocal microscopy, followed by in situ proximity ligation assay microscopy to confirm close-range (<16 nm) interaction.
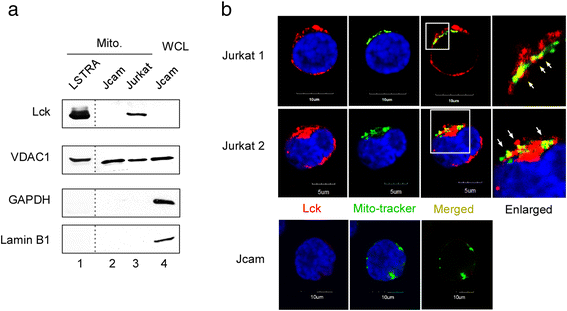
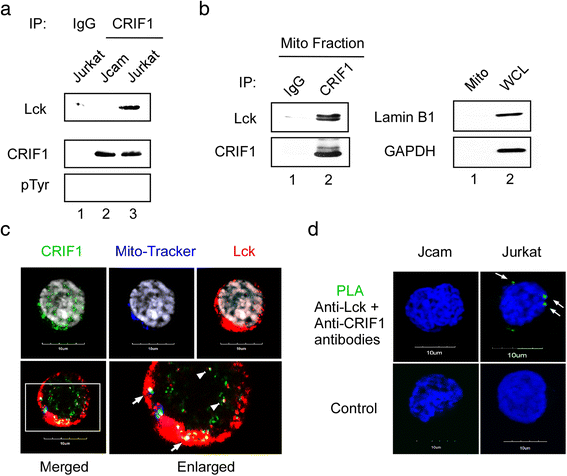
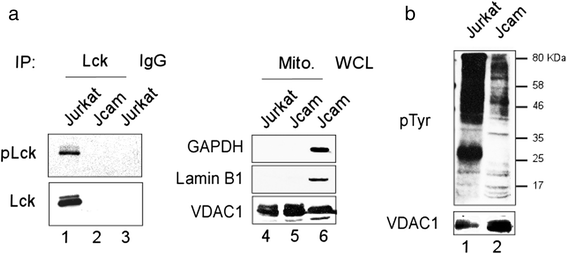
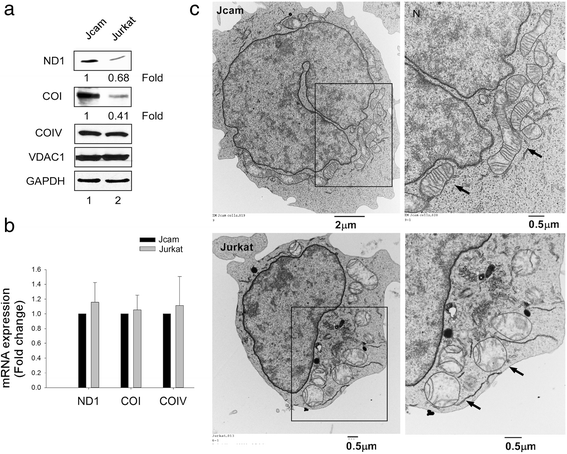
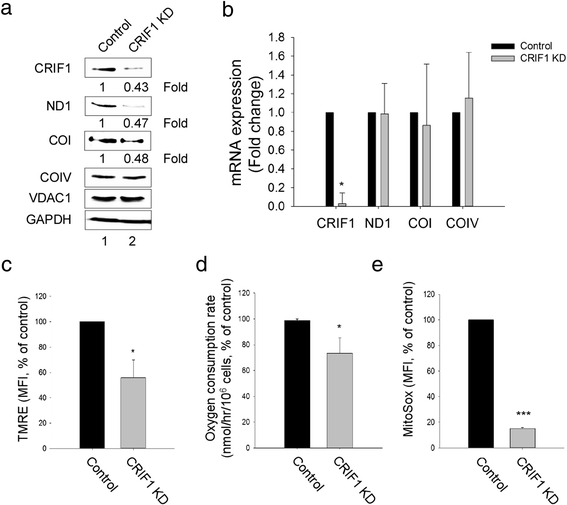
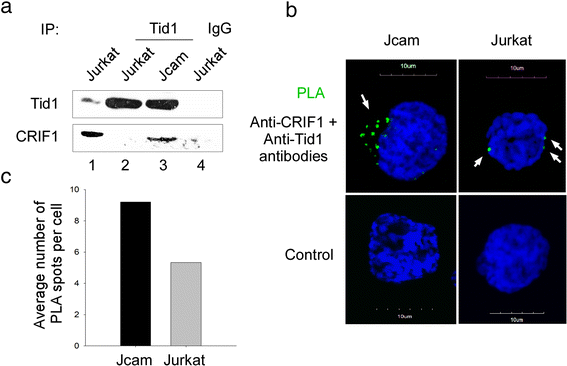
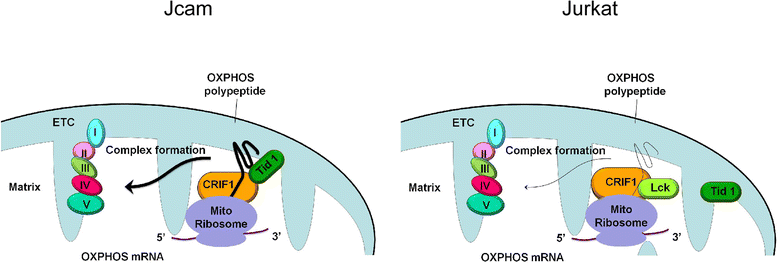
Results: Jurkat cells have abnormal mitochondrial structure and reduced levels of mitochondrial respiration, which is associated with the presence of mitochondrial Lck and lower levels of mitochondrion-encoded electron transport chain proteins. Proteomics identified CR6-interacting factor 1 (CRIF1) as the novel Lck-interacting protein. Lck association with CRIF1 in Jurkat mitochondria was confirmed biochemically and by microscopy, but did not lead to CRIF1 tyrosine phosphorylation. Consistent with the role of CRIF1 in functional mitoribosome, shRNA-mediated silencing of CRIF1 in Jcam resulted in mitochondrial dysfunction similar to that observed in Jurkat. Reduced interaction between CRIF1 and Tid1, another key component of intramitochondrial translational machinery, in Jurkat further supports the role of mitochondrial Lck as a negative regulator of CRIF1 through competitive binding.
Conclusions: This is the first report demonstrating the role of mitochondrial Lck in metabolic reprogramming of leukemic cells. Mechanistically, it is distinct from other reported mitochondrial protein tyrosine kinases. In a kinase-independent manner, mitochondrial Lck interferes with mitochondrial translational machinery through competitive binding to CRIF1. These findings may reveal novel approaches in cancer therapy by targeting cancer cell metabolism.
Figures
Similar articles
-
Nuclear lymphocyte-specific protein tyrosine kinase and its interaction with CR6-interacting factor 1 promote the survival of human leukemic T cells.Oncol Rep. 2015 Jul;34(1):43-50. doi: 10.3892/or.2015.3990. Epub 2015 May 19. Oncol Rep. 2015. PMID: 25997448 Free PMC article.
-
CR6-interacting factor 1 is a key regulator in Aβ-induced mitochondrial disruption and pathogenesis of Alzheimer's disease.Cell Death Differ. 2015 Jun;22(6):959-73. doi: 10.1038/cdd.2014.184. Epub 2014 Nov 7. Cell Death Differ. 2015. PMID: 25361083 Free PMC article.
-
CR6-Interacting Factor 1 Deficiency Impairs Vascular Function by Inhibiting the Sirt1-Endothelial Nitric Oxide Synthase Pathway.Antioxid Redox Signal. 2017 Aug 1;27(4):234-249. doi: 10.1089/ars.2016.6719. Epub 2017 May 24. Antioxid Redox Signal. 2017. PMID: 28117598
-
Multifunctions of CRIF1 in cancers and mitochondrial dysfunction.Front Oncol. 2022 Oct 3;12:1009948. doi: 10.3389/fonc.2022.1009948. eCollection 2022. Front Oncol. 2022. PMID: 36263222 Free PMC article. Review.
-
Mitoproteomics: Tackling Mitochondrial Dysfunction in Human Disease.Oxid Med Cell Longev. 2018 Nov 8;2018:1435934. doi: 10.1155/2018/1435934. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 30533169 Free PMC article. Review.
Cited by
-
The subtype-specific molecular function of SPDEF in breast cancer and insights into prognostic significance.J Cell Mol Med. 2021 Aug;25(15):7307-7320. doi: 10.1111/jcmm.16760. Epub 2021 Jun 30. J Cell Mol Med. 2021. PMID: 34191390 Free PMC article.
-
Co-Activation of PKC-δ by CRIF1 Modulates Oxidative Stress in Bone Marrow Multipotent Mesenchymal Stromal Cells after Irradiation by Phosphorylating NRF2 Ser40.Theranostics. 2017 Jun 25;7(10):2634-2648. doi: 10.7150/thno.17853. eCollection 2017. Theranostics. 2017. PMID: 28819452 Free PMC article.
-
Fatty Acid Oxidation-Driven Src Links Mitochondrial Energy Reprogramming and Oncogenic Properties in Triple-Negative Breast Cancer.Cell Rep. 2016 Mar 8;14(9):2154-2165. doi: 10.1016/j.celrep.2016.02.004. Epub 2016 Feb 25. Cell Rep. 2016. PMID: 26923594 Free PMC article.
-
CP-25 Attenuates the Activation of CD4+ T Cells Stimulated with Immunoglobulin D in Human.Front Pharmacol. 2018 Jan 23;9:4. doi: 10.3389/fphar.2018.00004. eCollection 2018. Front Pharmacol. 2018. PMID: 29410624 Free PMC article.
-
Identifying cancer tissue-of-origin by a novel machine learning method based on expression quantitative trait loci.Front Oncol. 2022 Aug 9;12:946552. doi: 10.3389/fonc.2022.946552. eCollection 2022. Front Oncol. 2022. PMID: 36016607 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

