Tetraspanin 3 Is Required for the Development and Propagation of Acute Myelogenous Leukemia
- PMID: 26212080
- PMCID: PMC4664079
- DOI: 10.1016/j.stem.2015.06.006
Tetraspanin 3 Is Required for the Development and Propagation of Acute Myelogenous Leukemia
Abstract
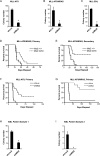
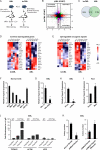
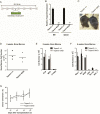
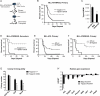
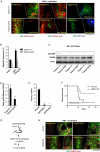
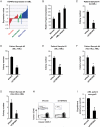
Acute Myelogenous Leukemia (AML) is an aggressive cancer that strikes both adults and children and is frequently resistant to therapy. Thus, identifying signals needed for AML propagation is a critical step toward developing new approaches for treating this disease. Here, we show that Tetraspanin 3 is a target of the RNA binding protein Musashi 2, which plays a key role in AML. We generated Tspan3 knockout mice that were born without overt defects. However, Tspan3 deletion impaired leukemia stem cell self-renewal and disease propagation and markedly improved survival in mouse models of AML. Additionally, Tspan3 inhibition blocked growth of AML patient samples, suggesting that Tspan3 is also important in human disease. As part of the mechanism, we show that Tspan3 deficiency disabled responses to CXCL12/SDF-1 and led to defects in AML localization within the niche. These identify Tspan3 as an important regulator of aggressive leukemias and highlight a role for Tspan3 in oncogenesis.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures
References
-
- Boucheix C, Duc GH, Jasmin C, Rubinstein E. Tetraspanins and malignancy. Expert reviews in molecular medicine. 2001;2001:1–17. - PubMed
-
- Byers RJ, Currie T, Tholouli E, Rodig SJ, Kutok JL. MSI2 protein expression predicts unfavorable outcome in acute myeloid leukemia. Blood. 2011;118:2857–2867. - PubMed
-
- Chen Y, Cortes J, Estrov Z, Faderl S, Qiao W, Abruzzo L, Garcia-Manero G, Pierce S, Huang X, Kebriaei P, et al. Persistence of cytogenetic abnormalities at complete remission after induction in patients with acute myeloid leukemia: prognostic significance and the potential role of allogeneic stem-cell transplantation. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:2507–2513. - PMC - PubMed
-
- Christiansen DH, Andersen MK, Desta F, Pedersen-Bjergaard J. Mutations of genes in the receptor tyrosine kinase (RTK)/RAS-BRAF signal transduction pathway in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2005;19:2232–2240. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- HL097767/HL/NHLBI NIH HHS/United States
- DP1 OD006430/OD/NIH HHS/United States
- DP1CA174422/CA/NCI NIH HHS/United States
- R01 CA186043/CA/NCI NIH HHS/United States
- DP1 CA174422/CA/NCI NIH HHS/United States
- CA186043/CA/NCI NIH HHS/United States
- R01 HL097767/HL/NHLBI NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- T32 GM007752/GM/NIGMS NIH HHS/United States
- P30CA23100/CA/NCI NIH HHS/United States
- R01 DK063031/DK/NIDDK NIH HHS/United States
- R01 DK099335/DK/NIDDK NIH HHS/United States
- DK099335/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
