Quantitative Proteomics Identifies Serum Response Factor Binding Protein 1 as a Host Factor for Hepatitis C Virus Entry
- PMID: 26212323
- PMCID: PMC4836839
- DOI: 10.1016/j.celrep.2015.06.063
Quantitative Proteomics Identifies Serum Response Factor Binding Protein 1 as a Host Factor for Hepatitis C Virus Entry
Abstract
Hepatitis C virus (HCV) enters human hepatocytes through a multistep mechanism involving, among other host proteins, the virus receptor CD81. How CD81 governs HCV entry is poorly characterized, and CD81 protein interactions after virus binding remain elusive. We have developed a quantitative proteomics protocol to identify HCV-triggered CD81 interactions and found 26 dynamic binding partners. At least six of these proteins promote HCV infection, as indicated by RNAi. We further characterized serum response factor binding protein 1 (SRFBP1), which is recruited to CD81 during HCV uptake and supports HCV infection in hepatoma cells and primary human hepatocytes. SRFBP1 facilitates host cell penetration by all seven HCV genotypes, but not of vesicular stomatitis virus and human coronavirus. Thus, SRFBP1 is an HCV-specific, pan-genotypic host entry factor. These results demonstrate the use of quantitative proteomics to elucidate pathogen entry and underscore the importance of host protein-protein interactions during HCV invasion.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
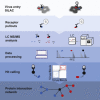
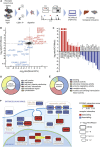
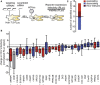
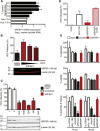

Comment in
-
SRFBP1, an Additional Player in HCV Entry.Trends Microbiol. 2015 Oct;23(10):590-593. doi: 10.1016/j.tim.2015.08.003. Epub 2015 Aug 26. Trends Microbiol. 2015. PMID: 26319372
Similar articles
-
Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB.PLoS Pathog. 2018 Jul 19;14(7):e1007111. doi: 10.1371/journal.ppat.1007111. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 30024968 Free PMC article.
-
SRFBP1, an Additional Player in HCV Entry.Trends Microbiol. 2015 Oct;23(10):590-593. doi: 10.1016/j.tim.2015.08.003. Epub 2015 Aug 26. Trends Microbiol. 2015. PMID: 26319372
-
Mice Expressing Minimally Humanized CD81 and Occludin Genes Support Hepatitis C Virus Uptake In Vivo.J Virol. 2017 Jan 31;91(4):e01799-16. doi: 10.1128/JVI.01799-16. Print 2017 Feb 15. J Virol. 2017. PMID: 27928007 Free PMC article.
-
CD81 and hepatitis C virus (HCV) infection.Viruses. 2014 Feb 6;6(2):535-72. doi: 10.3390/v6020535. Viruses. 2014. PMID: 24509809 Free PMC article. Review.
-
Hepatitis C virus entry: role of host and viral factors.Infect Genet Evol. 2012 Dec;12(8):1699-709. doi: 10.1016/j.meegid.2012.07.010. Epub 2012 Aug 2. Infect Genet Evol. 2012. PMID: 22878095 Review.
Cited by
-
New perspectives for preventing hepatitis C virus liver graft infection.Lancet Infect Dis. 2016 Jun;16(6):735-745. doi: 10.1016/S1473-3099(16)00120-1. Lancet Infect Dis. 2016. PMID: 27301929 Free PMC article. Review.
-
Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB.PLoS Pathog. 2018 Jul 19;14(7):e1007111. doi: 10.1371/journal.ppat.1007111. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 30024968 Free PMC article.
-
Decoding protein networks during virus entry by quantitative proteomics.Virus Res. 2016 Jun 15;218:25-39. doi: 10.1016/j.virusres.2015.09.006. Epub 2015 Sep 10. Virus Res. 2016. PMID: 26365680 Free PMC article. Review.
-
Whole-cell response of coronavirus-infected BMDCs through proteomic and transcriptomic analyses.Front Immunol. 2025 Jun 6;16:1513952. doi: 10.3389/fimmu.2025.1513952. eCollection 2025. Front Immunol. 2025. PMID: 40547011 Free PMC article.
-
Dynamic Dystroglycan Complexes Mediate Cell Entry of Lassa Virus.mBio. 2019 Mar 26;10(2):e02869-18. doi: 10.1128/mBio.02869-18. mBio. 2019. PMID: 30914516 Free PMC article.
References
-
- Benali-Furet N.L., Chami M., Houel L., De Giorgi F., Vernejoul F., Lagorce D., Buscail L., Bartenschlager R., Ichas F., Rizzuto R., Paterlini-Bréchot P. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene. 2005;24:4921–4933. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

