Doubling Throughput of a Real-Time PCR
- PMID: 26213283
- PMCID: PMC4515875
- DOI: 10.1038/srep12595
Doubling Throughput of a Real-Time PCR
Abstract
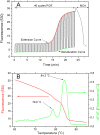
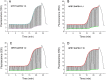
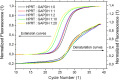
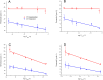
The invention of polymerase chain reaction (PCR) in 1983 revolutionized many areas of science, due to its ability to multiply a number of copies of DNA sequences (known as amplicons). Here we report on a method to double the throughput of quantitative PCR which could be especially useful for PCR-based mass screening. We concurrently amplified two target genes using only single fluorescent dye. A FAM probe labelled olionucleotide was attached to a quencher for one amplicon while the second one was without a probe. The PCR was performed in the presence of the intercalating dye SYBR Green I. We collected the fluorescence amplitude at two points per PCR cycle, at the denaturation and extension steps. The signal at denaturation is related only to the amplicon with the FAM probe while the amplitude at the extension contained information from both amplicons. We thus detected two genes within the same well using a single fluorescent channel. Any commercial real-time PCR systems can use this method doubling the number of detected genes. The method can be used for absolute quantification of DNA using a known concentration of housekeeping gene at one fluorescent channel.
Figures
Similar articles
-
[Quantitative PCR in the diagnosis of Leishmania].Parassitologia. 2004 Jun;46(1-2):163-7. Parassitologia. 2004. PMID: 15305709 Review. Italian.
-
Two-dye and one- or two-quencher DNA probes for real-time PCR assay: synthesis and comparison with a TaqMan™ probe.Anal Bioanal Chem. 2012 Jul;404(1):59-68. doi: 10.1007/s00216-012-6114-4. Epub 2012 Jun 19. Anal Bioanal Chem. 2012. PMID: 22710565
-
Single Fluorescence Channel-based Multiplex Detection of Avian Influenza Virus by Quantitative PCR with Intercalating Dye.Sci Rep. 2015 Jun 19;5:11479. doi: 10.1038/srep11479. Sci Rep. 2015. PMID: 26088868 Free PMC article.
-
Optimization of the Divergent method for genotyping single nucleotide variations using SYBR Green-based single-tube real-time PCR.Mutat Res. 2014 Aug-Sep;766-767:14-8. doi: 10.1016/j.mrfmmm.2014.05.013. Epub 2014 Jun 10. Mutat Res. 2014. PMID: 25847266
-
The real-time polymerase chain reaction.Mol Aspects Med. 2006 Apr-Jun;27(2-3):95-125. doi: 10.1016/j.mam.2005.12.007. Epub 2006 Feb 3. Mol Aspects Med. 2006. PMID: 16460794 Review.
Cited by
-
So Many Diagnostic Tests, So Little Time: Review and Preview of Candida auris Testing in Clinical and Public Health Laboratories.Front Microbiol. 2021 Oct 7;12:757835. doi: 10.3389/fmicb.2021.757835. eCollection 2021. Front Microbiol. 2021. PMID: 34691009 Free PMC article. Review.
-
Association of polymorphisms at the microRNA binding site of the caprine KITLG 3'-UTR with litter size.Sci Rep. 2016 May 11;6:25691. doi: 10.1038/srep25691. Sci Rep. 2016. PMID: 27168023 Free PMC article.
-
Significant Expansion of Real-Time PCR Multiplexing with Traditional Chemistries using Amplitude Modulation.Sci Rep. 2019 Jan 31;9(1):1053. doi: 10.1038/s41598-018-37732-y. Sci Rep. 2019. PMID: 30705333 Free PMC article.
-
IoT PCR for pandemic disease detection and its spread monitoring.Sens Actuators B Chem. 2020 Jan 15;303:127098. doi: 10.1016/j.snb.2019.127098. Epub 2019 Sep 11. Sens Actuators B Chem. 2020. PMID: 32288256 Free PMC article.
-
Molecular Diagnosis of Leishmaniasis: Quantification of Parasite Load by a Real-Time PCR Assay with High Sensitivity.Pathogens. 2021 Jul 9;10(7):865. doi: 10.3390/pathogens10070865. Pathogens. 2021. PMID: 34358015 Free PMC article.
References
-
- Saiki R. et al. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230, 1350–1354 (1985). - PubMed
-
- Higuchi R., Fockler C., Dollinger G. & Watson R. Kinetic PCR Analysis: Real-time Monitoring of DNA Amplification Reactions. Nat Biotech 11, 1026–1030 (1993). - PubMed
-
- Andrus A. et al. High-throughput synthesis of functionalized oligonucleotides. Nucleic acids symposium series, 317–318 (1997). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

