Quantifying selective pressures driving bacterial evolution using lineage analysis
- PMID: 26213639
- PMCID: PMC4511495
- DOI: 10.1103/PhysRevX.5.011016
Quantifying selective pressures driving bacterial evolution using lineage analysis
Abstract
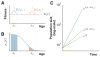
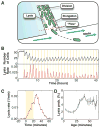
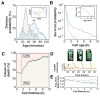
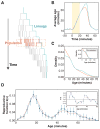
Organisms use a variety of strategies to adapt to their environments and maximize long-term growth potential, but quantitative characterization of the benefits conferred by the use of such strategies, as well as their impact on the whole population's rate of growth, remains challenging. Here, we use a path-integral framework that describes how selection acts on lineages -i.e. the life-histories of individuals and their ancestors- to demonstrate that lineage-based measurements can be used to quantify the selective pressures acting on a population. We apply this analysis to E. coli bacteria exposed to cyclical treatments of carbenicillin, an antibiotic that interferes with cell-wall synthesis and affects cells in an age-dependent manner. While the extensive characterization of the life-history of thousands of cells is necessary to accurately extract the age-dependent selective pressures caused by carbenicillin, the same measurement can be recapitulated using lineage-based statistics of a single surviving cell. Population-wide evolutionary pressures can be extracted from the properties of the surviving lineages within a population, providing an alternative and efficient procedure to quantify the evolutionary forces acting on a population. Importantly, this approach is not limited to age-dependent selection, and the framework can be generalized to detect signatures of other trait-specific selection using lineage-based measurements. Our results establish a powerful way to study the evolutionary dynamics of life under selection, and may be broadly useful in elucidating selective pressures driving the emergence of antibiotic resistance and the evolution of survival strategies in biological systems.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
