The Structural Determinants behind the Epigenetic Role of Histone Variants
- PMID: 26213973
- PMCID: PMC4584325
- DOI: 10.3390/genes6030685
The Structural Determinants behind the Epigenetic Role of Histone Variants
Abstract
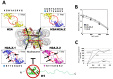
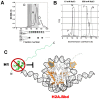
Histone variants are an important part of the histone contribution to chromatin epigenetics. In this review, we describe how the known structural differences of these variants from their canonical histone counterparts impart a chromatin signature ultimately responsible for their epigenetic contribution. In terms of the core histones, H2A histone variants are major players while H3 variant CenH3, with a controversial role in the nucleosome conformation, remains the genuine epigenetic histone variant. Linker histone variants (histone H1 family) haven't often been studied for their role in epigenetics. However, the micro-heterogeneity of the somatic canonical forms of linker histones appears to play an important role in maintaining the cell-differentiated states, while the cell cycle independent linker histone variants are involved in development. A picture starts to emerge in which histone H2A variants, in addition to their individual specific contributions to the nucleosome structure and dynamics, globally impair the accessibility of linker histones to defined chromatin locations and may have important consequences for determining different states of chromatin metabolism.
Keywords: chromatin; epigenetics; histone variants; histones; nucleosome.
Figures





References
-
- Kossel A. Uber einen peptonartigen bestandteil des zellkerns. Z. Physiol. Chem. 1884;8:511–515.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

