Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics
- PMID: 26214591
- PMCID: PMC4564951
- DOI: 10.1038/ng.3360
Analysis of mammalian gene function through broad-based phenotypic screens across a consortium of mouse clinics
Abstract
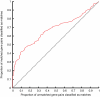
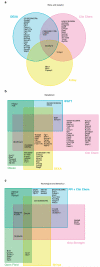
The function of the majority of genes in the mouse and human genomes remains unknown. The mouse embryonic stem cell knockout resource provides a basis for the characterization of relationships between genes and phenotypes. The EUMODIC consortium developed and validated robust methodologies for the broad-based phenotyping of knockouts through a pipeline comprising 20 disease-oriented platforms. We developed new statistical methods for pipeline design and data analysis aimed at detecting reproducible phenotypes with high power. We acquired phenotype data from 449 mutant alleles, representing 320 unique genes, of which half had no previous functional annotation. We captured data from over 27,000 mice, finding that 83% of the mutant lines are phenodeviant, with 65% demonstrating pleiotropy. Surprisingly, we found significant differences in phenotype annotation according to zygosity. New phenotypes were uncovered for many genes with previously unknown function, providing a powerful basis for hypothesis generation and further investigation in diverse systems.
Figures


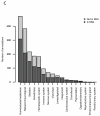
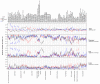
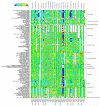
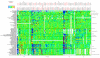
Comment in
-
Genetic differential calculus.Nat Genet. 2015 Sep;47(9):965-6. doi: 10.1038/ng.3384. Nat Genet. 2015. PMID: 26313224
-
Scaling up phenotyping studies.Nat Biotechnol. 2015 Nov;33(11):1150-1. doi: 10.1038/nbt.3400. Nat Biotechnol. 2015. PMID: 26544146 No abstract available.
References
-
- Brown SD, Wurst W, Kuhn R, Hancock JM. The functional annotation of mammalian genomes: the challenge of phenotyping. Annu Rev Genet. 2009;43:305–33. - PubMed
-
- Gailus-Durner V, et al. Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat Methods. 2005;2:403–4. - PubMed
-
- Wagner GP, Zhang J. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 2011;12:204–13. - PubMed
Additional References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

