Mouse genome annotation by the RefSeq project
- PMID: 26215545
- PMCID: PMC4602073
- DOI: 10.1007/s00335-015-9585-8
Mouse genome annotation by the RefSeq project
Abstract
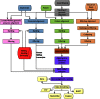
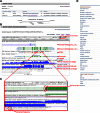
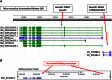
Complete and accurate annotation of the mouse genome is critical to the advancement of research conducted on this important model organism. The National Center for Biotechnology Information (NCBI) develops and maintains many useful resources to assist the mouse research community. In particular, the reference sequence (RefSeq) database provides high-quality annotation of multiple mouse genome assemblies using a combinatorial approach that leverages computation, manual curation, and collaboration. Implementation of this conservative and rigorous approach, which focuses on representation of only full-length and non-redundant data, produces high-quality annotation products. RefSeq records explicitly link sequences to current knowledge in a timely manner, updating public records regularly and rapidly in response to nomenclature updates, addition of new relevant publications, collaborator discussion, and user feedback. Whole genome re-annotation is also conducted at least every 12-18 months, and often more frequently in response to assembly updates or availability of informative data. This article highlights key features and advantages of RefSeq genome annotation products and presents an overview of NCBI processes to generate these data. Further discussion of NCBI's resources highlights useful features and the best methods for accessing our data.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

