Epiregulin contributes to breast tumorigenesis through regulating matrix metalloproteinase 1 and promoting cell survival
- PMID: 26215578
- PMCID: PMC4517352
- DOI: 10.1186/s12943-015-0408-z
Epiregulin contributes to breast tumorigenesis through regulating matrix metalloproteinase 1 and promoting cell survival
Abstract
Background: The epidermal growth factor (EGF) family of ligands has been implicated in promoting breast cancer initiation, growth and progression. The contributions of EGF family ligands and their receptors to breast cancer are complex, and the specific mechanisms through which different ligands regulate breast tumor initiation and growth are not well-defined. These studies focus on the EGF family member epiregulin (EREG) as a mediator of early stage breast tumorigenesis.
Methods: EREG expression levels were assessed in both cell lines and human samples of ductal carcinoma in situ (DCIS) using quantitative RT-PCR, ELISA and immunohistochemistry. Gene knock-down approaches using shRNA-based strategies were used to determine the requirement of EREG for growth of MCF10DCIS cells in vivo, and for identifying mechanisms through which EREG promotes tumor cell survival. Experiments were performed using a combination of two-dimensional culture, three-dimensional culture and tumor growth in vivo.
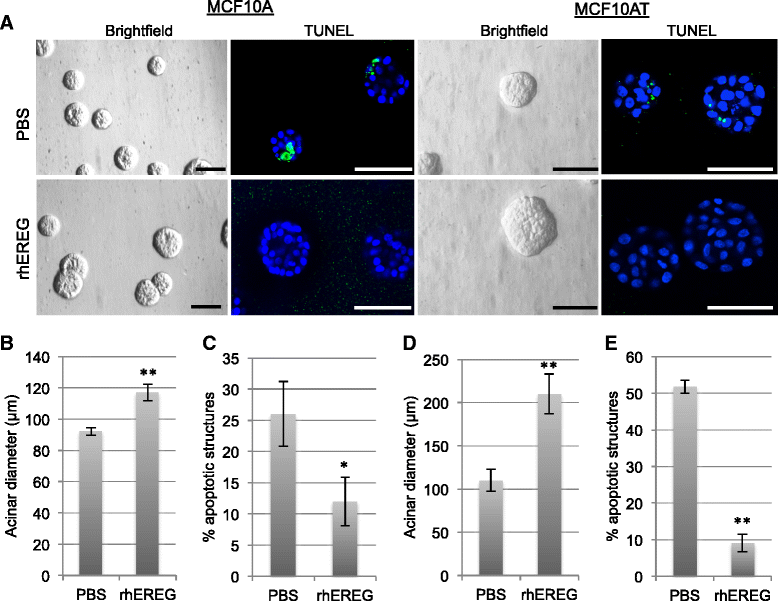
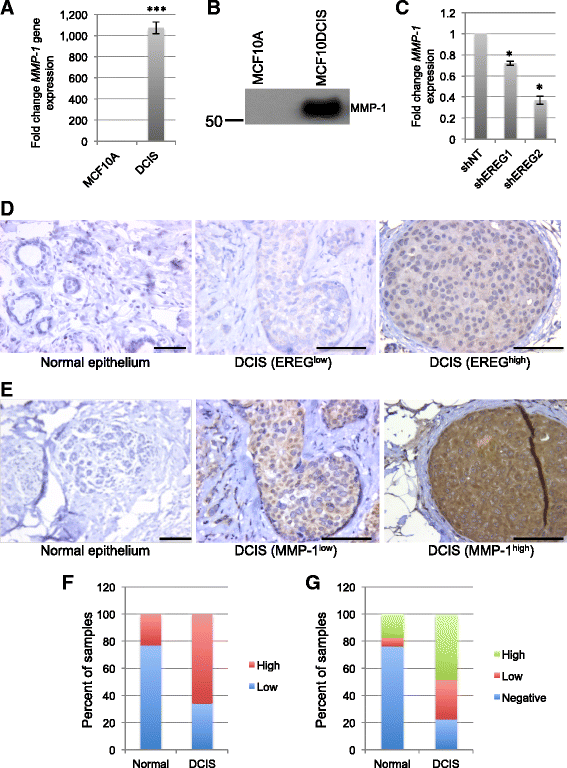
Results: In comparison with other EGF family members, EREG was induced in MCF10DCIS cells compared with MCF10A and MCF10AT cells and its expression was partially regulated by fibroblast growth factor receptor (FGFR) activity. Reduced EREG expression in MCF10DCIS cells led to decreased tumor growth in vivo, which was associated with reduced cell survival. Furthermore, treatment of MCF10A cells with exogenous EREG enhanced cell survival both in three-dimensional culture and in response to chemotherapeutic agents. Examination of EREG-induced signaling pathways demonstrated that EREG promoted survival of MCF10A cells through regulating expression of matrix metalloproteinase-1 (MMP-1). To determine the relevance of these findings in human tumors, samples of DCIS were analyzed for EREG and MMP-1 expression. EREG was induced in DCIS lesions compared to normal breast epithelium, and EREG and MMP-1 were correlated in a subset of DCIS samples.
Conclusions: Together, these studies lead to identification of a novel pathway involving EREG and MMP-1 that contributes to the formation of early stage breast cancer. Understanding these complex pathways could ultimately lead to the development of novel biomarkers of neoplastic progression and/or new therapeutic strategies for patients with early stage cancer.
Figures




References
-
- Boyle DP, Mullan P, Salto-Tellez M. Molecular mapping the presence of druggable targets in preinvasive and precursor breast lesions: a comprehensive review of biomarkers related to therapeutic interventions. Biochim Biophys Acta. 2013;1835(2):230–42. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

