Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers
- PMID: 26216839
- PMCID: PMC4589488
- DOI: 10.1053/j.gastro.2015.07.011
Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers
Abstract
Colorectal cancer (CRC) is a leading cause of cancer deaths worldwide. One of the fundamental processes driving the initiation and progression of CRC is the accumulation of a variety of genetic and epigenetic changes in colonic epithelial cells. Over the past decade, major advances have been made in our understanding of cancer epigenetics, particularly regarding aberrant DNA methylation, microRNA (miRNA) and noncoding RNA deregulation, and alterations in histone modification states. Assessment of the colon cancer "epigenome" has revealed that virtually all CRCs have aberrantly methylated genes and altered miRNA expression. The average CRC methylome has hundreds to thousands of abnormally methylated genes and dozens of altered miRNAs. As with gene mutations in the cancer genome, a subset of these epigenetic alterations, called driver events, are presumed to have a functional role in CRC. In addition, the advances in our understanding of epigenetic alterations in CRC have led to these alterations being developed as clinical biomarkers for diagnostic, prognostic, and therapeutic applications. Progress in this field suggests that these epigenetic alterations will be commonly used in the near future to direct the prevention and treatment of CRC.
Keywords: DNA Methylation; Histone Modification; Long Noncoding RNA; MicroRNA.
Copyright © 2015 AGA Institute. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures

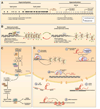
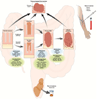
References
-
- Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61:212–236. - PubMed
-
- CH W. The epigenotype. Endeavour. 1942;1:18–20.
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. - PubMed
-
- Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA015704/CA/NCI NIH HHS/United States
- P01 CA077852/CA/NCI NIH HHS/United States
- R01 CA72851/CA/NCI NIH HHS/United States
- U54 CA143862/CA/NCI NIH HHS/United States
- CA181572/CA/NCI NIH HHS/United States
- U01 CA187956/CA/NCI NIH HHS/United States
- CA184792/CA/NCI NIH HHS/United States
- R01 CA181572/CA/NCI NIH HHS/United States
- R01 CA072851/CA/NCI NIH HHS/United States
- R01 CA115513/CA/NCI NIH HHS/United States
- U01 CA152756/CA/NCI NIH HHS/United States
- R01 CA194663/CA/NCI NIH HHS/United States
- R01 CA184792/CA/NCI NIH HHS/United States
- U01CA152756/CA/NCI NIH HHS/United States
- U54CA143862/CA/NCI NIH HHS/United States
- P01CA077852/CA/NCI NIH HHS/United States
- P30CA15704/CA/NCI NIH HHS/United States
- R01CA194663/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

