The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation
- PMID: 26216949
- PMCID: PMC4538674
- DOI: 10.1073/pnas.1502159112
The isolation of an RNA aptamer targeting to p53 protein with single amino acid mutation
Abstract
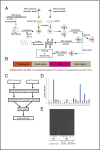
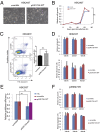
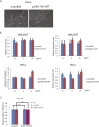
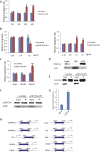
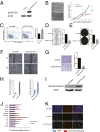
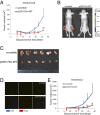
p53, known as a tumor suppressor, is a DNA binding protein that regulates cell cycle, activates DNA repair proteins, and triggers apoptosis in multicellular animals. More than 50% of human cancers contain a mutation or deletion of the p53 gene, and p53R175 is one of the hot spots of p53 mutation. Nucleic acid aptamers are short single-stranded oligonucleotides that are able to bind various targets, and they are typically isolated from an experimental procedure called systematic evolution of ligand exponential enrichment (SELEX). Using a previously unidentified strategy of contrast screening with SELEX, we have isolated an RNA aptamer targeting p53R175H. This RNA aptamer (p53R175H-APT) has a significantly stronger affinity to p53R175H than to the wild-type p53 in both in vitro and in vivo assays. p53R175H-APT decreased the growth rate, weakened the migration capability, and triggered apoptosis in human lung cancer cells harboring p53R175H. Further analysis actually indicated that p53R175H-APT might partially rescue or correct the p53R175H to function more like the wild-type p53. In situ injections of p53R175H-APT to the tumor xenografts confirmed the effects of this RNA aptamer on p53R175H mutation in mice.
Keywords: RNA aptamer; SELEX; contrast screening; p53; tumor.
Conflict of interest statement
Conflict of interest statement: G.S. and L.C. have an ownership interest in a patent related to this research.
Figures


References
-
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. - PubMed
-
- Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. - PubMed
-
- Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355(6360):564–566. - PubMed
-
- Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34(2):656–665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

